Adeno-associated virus site-specific integration and AAVS1 disruption
- PMID: 15254160
- PMCID: PMC446113
- DOI: 10.1128/JVI.78.15.7874-7882.2004
Adeno-associated virus site-specific integration and AAVS1 disruption
Abstract
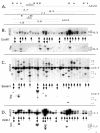
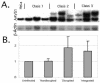
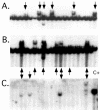
Adeno-associated virus (AAV) is a single-stranded DNA virus with a unique biphasic lifestyle consisting of both a productive and a latent phase. Typically, the productive phase requires coinfection with a helper virus, for instance adenovirus, while the latent phase dominates in healthy cells. In the latent state, AAV is found integrated site specifically into the host genome at chromosome 19q13.4 qtr (AAVS1), the only animal virus known to integrate in a defined location. In this study we investigated the latent phase of serotype 2 AAV, focusing on three areas: AAV infection, rescue, and integration efficiency as a function of viral multiplicity of infection (MOI); efficiency of site-specific integration; and disruption of the AAVS1 locus. As expected, increasing the AAV MOI resulted in an increase in the percentage of cells infected, with 80% of cells infected at an MOI of 10. Additional MOI only marginally effected a further increase in percentage of infected cells. In contrast to infection, we found very low levels of integration at MOIs of less than 10. At an MOI of 10, at which 80% of cells are infected, less than 5% of clonal cell lines contained integrated AAV DNA. At an MOI of 100 or greater, however, 35 to 40% of clonal cell lines contained integrated AAV DNA. Integration and the ability to rescue viral genomes were highly correlated. Analysis of integrated AAV indicated that essentially all integrants were AAVS1 site specific. Although maximal integration efficiency approached 40% of clonal cell lines (essentially 50% of infected cells), over 80% of cell lines contained a genomic disruption at the AAVS1 integration locus on chromosome 19 ( approximately 100% of infected cells). Rep expression by itself and in the presence of a plasmid integration substrate was able to mediate this disruption of the AAVS1 site. We further characterized the disruption event and demonstrated that it resulted in amplification of the AAVS1 locus. The data are consistent with a revised model of AAV integration that includes preliminary expansion of a defined region in AAVS1.
Figures
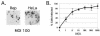


References
-
- Berns, K. I., T. C. Pinkerton, G. F. Thomas, and M. D. Hoggan. 1975. Detection of adeno-associated virus (AAV)-specific nucleotide sequences in DNA isolated from latently infected Detroit 6 cells. Virology 68:556-560. - PubMed
-
- Berns, K. I., and R. M. Linden. 1995. The cryptic life style of adeno-associated virus. Bioessays 17:237-245. - PubMed
-
- Berns, K. I. 1996. Parvoviridae: the viruses and their replication, p. 1017-1041. In B. N. Fields (ed.), Fundamental virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia Pa.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

