Comprehensive mutational analysis of a herpesvirus gene in the viral genome context reveals a region essential for virus replication
- PMID: 15254174
- PMCID: PMC446129
- DOI: 10.1128/JVI.78.15.8026-8035.2004
Comprehensive mutational analysis of a herpesvirus gene in the viral genome context reveals a region essential for virus replication
Abstract
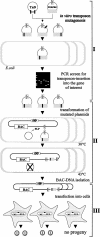
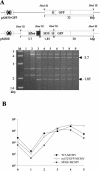
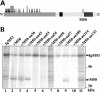
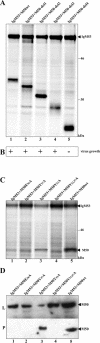
Essential viral proteins perform vital functions during morphogenesis via a complex interaction with other viral and cellular gene products. Here, we present a novel approach to comprehensive mutagenesis of essential cytomegalovirus genes and biological analysis in the 230-kbp-genome context. A random Tn7-based mutagenesis procedure at the single-gene level was combined with site-specific recombination via the FLP/FLP recognition target site system for viral genome reconstitution. We show the function of more than 100 mutants from a larger library of M50/p35, a protein involved in capsid egress from the nucleus. This protein recruits other viral proteins and cellular enzymes to the inner nuclear membrane. Our approach enabled us to rapidly discriminate between essential and nonessential regions within the coding sequence. Based on the prediction of the screen, we were able to map a site essential for viral protein-protein interaction at the amino acid level.
Figures

Similar articles
-
Structure of a herpesvirus nuclear egress complex subunit reveals an interaction groove that is essential for viral replication.Proc Natl Acad Sci U S A. 2015 Jul 21;112(29):9010-5. doi: 10.1073/pnas.1511140112. Epub 2015 Jul 6. Proc Natl Acad Sci U S A. 2015. PMID: 26150520 Free PMC article.
-
Random screening for dominant-negative mutants of the cytomegalovirus nuclear egress protein M50.J Virol. 2007 Jun;81(11):5508-17. doi: 10.1128/JVI.02796-06. Epub 2007 Mar 21. J Virol. 2007. PMID: 17376929 Free PMC article.
-
Functional domains of murine cytomegalovirus nuclear egress protein M53/p38.J Virol. 2006 Jan;80(1):73-84. doi: 10.1128/JVI.80.1.73-84.2006. J Virol. 2006. PMID: 16352532 Free PMC article.
-
A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism.Science. 2001 Jan 12;291(5502):303-5. doi: 10.1126/science.291.5502.303. Science. 2001. PMID: 11209080
-
Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina.Science. 2002 Aug 2;297(5582):854-7. doi: 10.1126/science.1071506. Science. 2002. PMID: 12161659
Cited by
-
Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B.J Virol. 2006 Jan;80(1):494-504. doi: 10.1128/JVI.80.1.494-504.2006. J Virol. 2006. PMID: 16352573 Free PMC article.
-
The Complex Regulatory Role of Cytomegalovirus Nuclear Egress Protein pUL50 in the Production of Infectious Virus.Cells. 2021 Nov 11;10(11):3119. doi: 10.3390/cells10113119. Cells. 2021. PMID: 34831342 Free PMC article.
-
Evolutionarily conserved herpesviral protein interaction networks.PLoS Pathog. 2009 Sep;5(9):e1000570. doi: 10.1371/journal.ppat.1000570. Epub 2009 Sep 4. PLoS Pathog. 2009. PMID: 19730696 Free PMC article.
-
Characterization of conserved region 2-deficient mutants of the cytomegalovirus egress protein pM53.J Virol. 2012 Dec;86(23):12512-24. doi: 10.1128/JVI.00471-12. Epub 2012 Sep 19. J Virol. 2012. PMID: 22993161 Free PMC article.
-
Structure of a herpesvirus nuclear egress complex subunit reveals an interaction groove that is essential for viral replication.Proc Natl Acad Sci U S A. 2015 Jul 21;112(29):9010-5. doi: 10.1073/pnas.1511140112. Epub 2015 Jul 6. Proc Natl Acad Sci U S A. 2015. PMID: 26150520 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

