Sequential roles of receptor binding and low pH in forming prehairpin and hairpin conformations of a retroviral envelope glycoprotein
- PMID: 15254191
- PMCID: PMC446138
- DOI: 10.1128/JVI.78.15.8201-8209.2004
Sequential roles of receptor binding and low pH in forming prehairpin and hairpin conformations of a retroviral envelope glycoprotein
Abstract
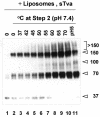
A general model has been proposed for the fusion mechanisms of class I viral fusion proteins. According to this model a metastable trimer, anchored in the viral membrane through its transmembrane domain, transits to a trimeric prehairpin intermediate, anchored at its opposite end in the target membrane through its fusion peptide. A subsequent refolding event creates a trimer of hairpins (often termed a six-helix bundle) in which the previously well-separated transmembrane domain and fusion peptide (and their attached membranes) are brought together, thereby driving membrane fusion. While there is ample biochemical and structural information on the trimer-of-hairpins conformation of class I viral fusion proteins, less is known about intermediate states between native metastable trimers and the final trimer of hairpins. In this study we analyzed conformational states of the transmembrane subunit (TM), the fusion subunit, of the Env glycoprotein of the subtype A avian sarcoma and leukosis virus (ASLV-A). By analyzing forms of EnvA TM on mildly denaturing sodium dodecyl sulfate gels we identified five conformational states of EnvA TM. Following interaction of virions with a soluble form of the ASLV-A receptor at 37 degrees C, the metastable form of EnvA TM (which migrates at 37 kDa) transits to a 70-kDa and then to a 150-kDa species. Following subsequent exposure to a low pH (or an elevated temperature or the fusion promoting agent chlorpromazine), an additional set of bands at >150 kDa, and then a final band at 100 kDa, forms. Both an EnvA C-helix peptide (which inhibits virus fusion and infectivity) and the fusion-inhibitory agent lysophosphatidylcholine inhibit the formation of the >150- and 100-kDa bands. Our data are consistent with the 70- and 150-kDa bands representing precursor and fully formed prehairpin conformations of EnvA TM. Our data are also consistent with the >150-kDa bands representing higher-order oligomers of EnvA TM and with the 100-kDa band representing the fully formed six-helix bundle. In addition to resolving fusion-relevant conformational intermediates of EnvA TM, our data are compatible with a model in which the EnvA protein is activated by its receptor (at neutral pH and a temperature greater than or equal to room temperature) to form prehairpin conformations of EnvA TM, and in which subsequent exposure to a low pH is required to stabilize the final six-helix bundle, which drives a later stage of fusion.
Figures




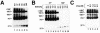
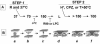
Similar articles
-
The avian retrovirus avian sarcoma/leukosis virus subtype A reaches the lipid mixing stage of fusion at neutral pH.J Virol. 2003 Mar;77(5):3058-66. doi: 10.1128/jvi.77.5.3058-3066.2003. J Virol. 2003. PMID: 12584331 Free PMC article.
-
Studies of the "chain reversal regions" of the avian sarcoma/leukosis virus (ASLV) and ebolavirus fusion proteins: analogous residues are important, and a His residue unique to EnvA affects the pH dependence of ASLV entry.J Virol. 2010 Jun;84(11):5687-94. doi: 10.1128/JVI.02583-09. Epub 2010 Mar 24. J Virol. 2010. PMID: 20335266 Free PMC article.
-
Receptor-induced conformational changes in the SU subunit of the avian sarcoma/leukosis virus A envelope protein: implications for fusion activation.J Virol. 2005 Mar;79(6):3488-99. doi: 10.1128/JVI.79.6.3488-3499.2005. J Virol. 2005. PMID: 15731243 Free PMC article.
-
Alpharetrovirus envelope-receptor interactions.Curr Top Microbiol Immunol. 2003;281:107-36. doi: 10.1007/978-3-642-19012-4_3. Curr Top Microbiol Immunol. 2003. PMID: 12932076 Review.
-
Cellular entry of retroviruses.Adv Exp Med Biol. 2013;790:128-49. doi: 10.1007/978-1-4614-7651-1_7. Adv Exp Med Biol. 2013. PMID: 23884589 Review.
Cited by
-
Structure-Based Mutations in the Herpes Simplex Virus 1 Glycoprotein B Ectodomain Arm Impart a Slow-Entry Phenotype.mBio. 2017 May 16;8(3):e00614-17. doi: 10.1128/mBio.00614-17. mBio. 2017. PMID: 28512095 Free PMC article.
-
Role of electrostatic repulsion in controlling pH-dependent conformational changes of viral fusion proteins.Structure. 2013 Jul 2;21(7):1085-96. doi: 10.1016/j.str.2013.05.009. Structure. 2013. PMID: 23823327 Free PMC article. Review.
-
Kinetic analyses of the surface-transmembrane disulfide bond isomerization-controlled fusion activation pathway in Moloney murine leukemia virus.J Virol. 2005 Nov;79(22):13856-64. doi: 10.1128/JVI.79.22.13856-13864.2005. J Virol. 2005. PMID: 16254321 Free PMC article.
-
Structural characterization of a fusion glycoprotein from a retrovirus that undergoes a hybrid 2-step entry mechanism.FASEB J. 2013 Dec;27(12):5059-71. doi: 10.1096/fj.13-232371. Epub 2013 Sep 13. FASEB J. 2013. PMID: 24036886 Free PMC article.
-
Cysteines flanking the internal fusion peptide are required for the avian sarcoma/leukosis virus glycoprotein to mediate the lipid mixing stage of fusion with high efficiency.J Virol. 2008 Mar;82(6):3131-4. doi: 10.1128/JVI.02266-07. Epub 2008 Jan 9. J Virol. 2008. PMID: 18184714 Free PMC article.
References
-
- Bates, P., J. A. T. Young, and H. E. Varmus. 1993. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell 74:1043-1051. - PubMed
-
- Bobkova, M., J. Stitz, M. Engelstadter, K. Cichutek, and C. J. Buchholz. 2002. Identification of R-peptides in envelope proteins of C-type retroviruses. J. Gen. Virol. 83:2241-2246. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

