Functional complementation of human centromere protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae
- PMID: 15254229
- PMCID: PMC444843
- DOI: 10.1128/MCB.24.15.6620-6630.2004
Functional complementation of human centromere protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae
Abstract
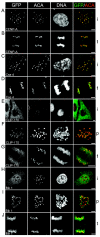
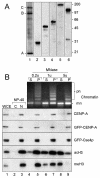
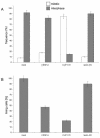
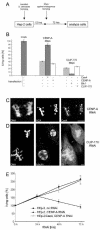
We have employed a novel in vivo approach to study the structure and function of the eukaryotic kinetochore multiprotein complex. RNA interference (RNAi) was used to block the synthesis of centromere protein A (CENP-A) and Clip-170 in human cells. By coexpression, homologous kinetochore proteins from Saccharomyces cerevisiae were then tested for the ability to complement the RNAi-induced phenotypes. Cse4p, the budding yeast CENP-A homolog, was specifically incorporated into kinetochore nucleosomes and was able to complement RNAi-induced cell cycle arrest in CENP-A-depleted human cells. Thus, Cse4p can structurally and functionally substitute for CENP-A, strongly suggesting that the basic features of centromeric chromatin are conserved between yeast and mammals. Bik1p, the budding yeast homolog of human CLIP-170, also specifically localized to kinetochores during mitosis, but Bik1p did not rescue CLIP-170 depletion-induced cell cycle arrest. Generally, the newly developed in vivo complementation assay provides a powerful new tool for studying the function and evolutionary conservation of multiprotein complexes from yeast to humans.
Figures

Similar articles
-
Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway.J Cell Biol. 2003 Jan 6;160(1):25-39. doi: 10.1083/jcb.200210005. Epub 2003 Jan 6. J Cell Biol. 2003. PMID: 12515822 Free PMC article.
-
Functional roles for evolutionarily conserved Spt4p at centromeres and heterochromatin in Saccharomyces cerevisiae.EMBO J. 2004 Apr 21;23(8):1804-14. doi: 10.1038/sj.emboj.7600161. Epub 2004 Apr 1. EMBO J. 2004. PMID: 15057281 Free PMC article.
-
The centromere-specific histone variant Cse4p (CENP-A) is essential for functional chromatin architecture at the yeast 2-microm circle partitioning locus and promotes equal plasmid segregation.J Cell Biol. 2006 Sep 11;174(6):779-90. doi: 10.1083/jcb.200603042. J Cell Biol. 2006. PMID: 16966420 Free PMC article.
-
How to build a centromere: from centromeric and pericentromeric chromatin to kinetochore assembly.Biochem Cell Biol. 2006 Aug;84(4):619-39. doi: 10.1139/o06-078. Biochem Cell Biol. 2006. PMID: 16936833 Review.
-
[DNA-binding proteins are involved in establishment of centromere chromatin structure coordinately with centromere specific histone variant, CENP-A].Tanpakushitsu Kakusan Koso. 2009 Aug;54(10):1276-83. Tanpakushitsu Kakusan Koso. 2009. PMID: 19663255 Review. Japanese. No abstract available.
Cited by
-
Co-evolving CENP-A and CAL1 Domains Mediate Centromeric CENP-A Deposition across Drosophila Species.Dev Cell. 2016 Apr 18;37(2):136-47. doi: 10.1016/j.devcel.2016.03.021. Dev Cell. 2016. PMID: 27093083 Free PMC article.
-
Transposons play an important role in the evolution and diversification of centromeres among closely related species.Front Plant Sci. 2015 Apr 7;6:216. doi: 10.3389/fpls.2015.00216. eCollection 2015. Front Plant Sci. 2015. PMID: 25904926 Free PMC article.
-
Heterochromatin and RNAi regulate centromeres by protecting CENP-A from ubiquitin-mediated degradation.PLoS Genet. 2018 Aug 8;14(8):e1007572. doi: 10.1371/journal.pgen.1007572. eCollection 2018 Aug. PLoS Genet. 2018. PMID: 30089114 Free PMC article.
-
The ABCs of CENPs.Chromosoma. 2011 Oct;120(5):425-46. doi: 10.1007/s00412-011-0330-0. Epub 2011 Jul 13. Chromosoma. 2011. PMID: 21751032 Review.
-
Centromere histone H3- and phospholipase-mediated haploid induction in plants.Plant Methods. 2019 Apr 26;15:42. doi: 10.1186/s13007-019-0429-5. eCollection 2019. Plant Methods. 2019. PMID: 31057661 Free PMC article. Review.
References
-
- Albertson, D. G., and J. N. Thomson. 1982. The kinetochores of Caenorhabditis elegans. Chromosoma 86:409-428. - PubMed
-
- Albertson, D. G., and J. N. Thomson. 1993. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1:15-26. - PubMed
-
- Amon, A. 1999. The spindle checkpoint. Curr. Opin. Genet. Dev. 9:69-75. - PubMed
-
- Biggins, S., and A. W. Murray. 1999. Sister chromatid cohesion in mitosis. Curr. Opin. Genet. Dev. 9:230-236. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
