Gene-targeted mice lacking the Trex1 (DNase III) 3'-->5' DNA exonuclease develop inflammatory myocarditis
- PMID: 15254239
- PMCID: PMC444847
- DOI: 10.1128/MCB.24.15.6719-6727.2004
Gene-targeted mice lacking the Trex1 (DNase III) 3'-->5' DNA exonuclease develop inflammatory myocarditis
Abstract
TREX1, originally designated DNase III, was isolated as a major nuclear DNA-specific 3'-->5' exonuclease that is widely distributed in both proliferating and nonproliferating mammalian tissues. The cognate cDNA shows homology to the editing subunit of the Escherichia coli replicative DNA polymerase III holoenzyme and encodes an exonuclease which was able to serve a DNA-editing function in vitro, promoting rejoining of a 3' mismatched residue in a reconstituted DNA base excision repair system. Here we report the generation of gene-targeted Trex1(-/-) mice. The null mice are viable and do not show the increase in spontaneous mutation frequency or cancer incidence that would be predicted if Trex1 served an obligatory role of editing mismatched 3' termini generated during DNA repair or DNA replication in vivo. Unexpectedly, Trex1(-/-) mice exhibit a dramatically reduced survival and develop inflammatory myocarditis leading to progressive, often dilated, cardiomyopathy and circulatory failure.
Figures
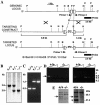
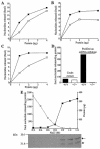
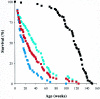
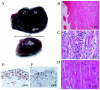

References
-
- Bachmaier, K., J. Le, and J. M. Penninger. 2000. “Catching heart disease”: antigenic mimicry and bacterial infections. Nat. Med. 6:841-842. - PubMed
-
- Brown, K. R., K. L. Weatherdon, C. L. Galligan, and V. Skalski. 2002. A nuclear 3′-5′ exonuclease proofreads for the exonuclease-deficient DNA polymerase α. DNA Repair 1:795-810. - PubMed
-
- Chou, K.-M., and Y.-C. Cheng. 2002. An exonucleolytic activity of human apurinic/apyrimidinic endonuclease on 3′ mispaired DNA. Nature 415:655-659. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
