A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p
- PMID: 15254252
- PMCID: PMC444856
- DOI: 10.1128/MCB.24.15.6871-6886.2004
A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p
Abstract
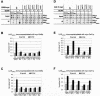
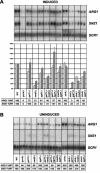
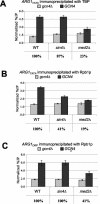
The Srb mediator is an important transcriptional coactivator for Gcn4p in the yeast Saccharomyces cerevisiae. We show that three subunits of the Gal11/tail domain of mediator, Gal11p, Pgd1p, and Med2p, and the head domain subunit Srb2p make overlapping contributions to the interaction of mediator with recombinant Gcn4p in vitro. Each of these proteins, along with the tail subunit Sin4p, also contributes to the recruitment of mediator by Gcn4p to target promoters in vivo. We found that Gal11p, Med2p, and Pgd1p reside in a stable subcomplex in sin4Delta cells that interacts with Gcn4p in vitro and that is recruited independently of the rest of mediator by Gcn4p in vivo. Thus, the Gal11p/Med2p/Pgd1p triad is both necessary for recruitment of intact mediator and appears to be sufficient for recruitment by Gcn4p as a free subcomplex. The med2Delta mutation impairs the recruitment of TATA binding protein (TBP) and RNA polymerase II to the promoter and the induction of transcription at ARG1, demonstrating the importance of the tail domain for activation by Gcn4p in vivo. Even though the Gal11p/Med2p/Pgd1p triad is the only portion of Srb mediator recruited efficiently to the promoter in the sin4Delta strain, this mutant shows high-level TBP recruitment and wild-type transcriptional induction at ARG1. Hence, the Gal11p/Med2p/Pgd1p triad may contribute to TBP recruitment independently of the rest of mediator.
Figures




References
-
- Brown, C., L. Howe, K. Sousa, S. C. Alley, M. J. Carrozza, S. Tan, and J. L. Workman. 2001. Recruitment of HAT complexes by direct activator interactions with the AATM-related Tra1 subunit. Science 292:2333-2337. - PubMed
-
- Chung, W. H., J. L. Craighead, W. H. Chang, C. Ezeokonkwo, A. Bareket-Samish, R. D. Kornberg, and F. J. Asturias. 2003. RNA polymerase II/TFIIF structure and conserved organization of the initiation complex. Mol. Cell 12:1003-1013. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
