Prevalence of intron gain over intron loss in the evolution of paralogous gene families
- PMID: 15254274
- PMCID: PMC484173
- DOI: 10.1093/nar/gkh686
Prevalence of intron gain over intron loss in the evolution of paralogous gene families
Abstract
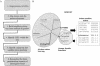
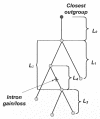
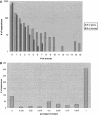
The mechanisms and evolutionary dynamics of intron insertion and loss in eukaryotic genes remain poorly understood. Reconstruction of parsimonious scenarios of gene structure evolution in paralogous gene families in animals and plants revealed numerous gains and losses of introns. In all analyzed lineages, the number of acquired new introns was substantially greater than the number of lost ancestral introns. This trend held even for lineages in which vertical evolution of genes involved more intron losses than gains, suggesting that gene duplication boosts intron insertion. However, dating gene duplications and the associated intron gains and losses based on the molecular clock assumption showed that very few, if any, introns were gained during the last approximately 100 million years of animal and plant evolution, in agreement with previous conclusions reached through analysis of orthologous gene sets. These results are generally compatible with the emerging notion of intensive insertion and loss of introns during transitional epochs in contrast to the relative quiet of the intervening evolutionary spans.
Figures



References
-
- Logsdon J.M. Jr (1998) The recent origins of spliceosomal introns revisited. Curr. Opin. Genet. Dev., 8, 637–648. - PubMed
-
- Lamond A.I. (1999) RNA splicing. Running rings around RNA. Nature, 397, 655–656. - PubMed
-
- Dacks J.B. and Doolittle,W.F. (2001) Reconstructing/deconstructing the earliest eukaryotes: how comparative genomics can help. Cell, 107, 419–425. - PubMed
-
- Lynch M. and Richardson,A.O. (2002) The evolution of spliceosomal introns. Curr. Opin. Genet. Dev., 12, 701–710. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

