Transcriptional regulation of the Drosophila caudal homeobox gene by DRE/DREF
- PMID: 15254275
- PMCID: PMC484175
- DOI: 10.1093/nar/gkh688
Transcriptional regulation of the Drosophila caudal homeobox gene by DRE/DREF
Abstract
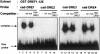
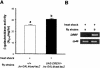
The caudal-related homeobox transcription factors are required for the normal development and differentiation of intestinal cells. Recent reports indicate that misregulation of homeotic gene expression is associated with gastrointestinal cancer in mammals. However, the molecular mechanisms that regulate expression of the caudal-related homeobox genes are poorly understood. In this study, we have identified a DNA replication-related element (DRE) in the 5' flanking region of the Drosophila caudal gene. Gel-mobility shift analysis reveals that three of the four DRE-related sequences in the caudal 5'-flanking region are recognized by the DRE-binding factor (DREF). Deletion and site-directed mutagenesis of these DRE sites results in a considerable reduction in caudal gene promoter activity. Analyses with transgenic flies carrying a caudal-lacZ fusion gene bearing wild-type or mutant DRE sites indicate that the DRE sites are required for caudal expression in vivo. These findings indicate that DRE/DREF is a key regulator of Drosophila caudal homeobox gene expression and suggest that DREs and DREF contribute to intestinal development by regulating caudal gene expression.
Figures

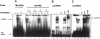




Similar articles
-
Transcriptional regulation of the Drosophila ANT gene by the DRE/DREF system.Genes Cells. 2007 May;12(5):569-79. doi: 10.1111/j.1365-2443.2007.01075.x. Genes Cells. 2007. PMID: 17535248
-
Transcriptional regulation of the Drosophila catalase gene by the DRE/DREF system.Nucleic Acids Res. 2004 Feb 24;32(4):1318-24. doi: 10.1093/nar/gkh302. Print 2004. Nucleic Acids Res. 2004. PMID: 14982956 Free PMC article.
-
Transcription control of a gene for Drosophila transcription factor, DREF by DRE and cis-elements conserved between Drosophila melanogaster and virilis.Gene. 2003 May 8;309(2):101-16. doi: 10.1016/s0378-1119(03)00493-1. Gene. 2003. PMID: 12758126
-
The DRE/DREF transcriptional regulatory system: a master key for cell proliferation.Biochim Biophys Acta. 2008 Feb;1779(2):81-9. doi: 10.1016/j.bbagrm.2007.11.011. Epub 2007 Dec 4. Biochim Biophys Acta. 2008. PMID: 18155677 Review.
-
Cooperativity in transcriptional control.Curr Biol. 2001 Apr 3;11(7):R250-2. doi: 10.1016/s0960-9822(01)00130-0. Curr Biol. 2001. PMID: 11413011 Review. No abstract available.
Cited by
-
Aging-related upregulation of the homeobox gene caudal represses intestinal stem cell differentiation in Drosophila.PLoS Genet. 2021 Jul 6;17(7):e1009649. doi: 10.1371/journal.pgen.1009649. eCollection 2021 Jul. PLoS Genet. 2021. PMID: 34228720 Free PMC article.
-
Genetic screening for modifiers of the DREF pathway in Drosophila melanogaster: identification and characterization of HP6 as a novel target of DREF.Nucleic Acids Res. 2009 Apr;37(5):1423-37. doi: 10.1093/nar/gkn1068. Epub 2009 Jan 9. Nucleic Acids Res. 2009. PMID: 19136464 Free PMC article.
References
-
- McGinnis W. and Krumlauf,R. (1992) Homeobox genes and axial patterning. Cell, 68, 283–302. - PubMed
-
- Dearolf C.R., Topol,J. and Parker,C.S. (1989) The caudal gene product is a direct activator of fushi tarazu transcription during Drosophila embyogenesis. Nature, 341, 340–343. - PubMed
-
- Mlodzik M., Gibson,G. and Gehring,W.J. (1990) Effects of ectopic expression of caudal during Drosophila development. Development, 109, 271–277. - PubMed
-
- Macdonald P.M. and Struhl,G. (1986) A molecular gradient in early Drosophila embryos and its role in specifying the body pattern. Nature, 324, 537–545. - PubMed
-
- Mlodzik M. and Gehring,W.J. (1987) Expression of the caudal gene in the germ line of Drosophila: formation of an RNA and protein gradient during early embryogenesis. Cell, 48, 465–478. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

