Human cytomegalovirus infection of tumor cells downregulates NCAM (CD56): a novel mechanism for virus-induced tumor invasiveness
- PMID: 15256054
- PMCID: PMC1502118
- DOI: 10.1593/neo.03418
Human cytomegalovirus infection of tumor cells downregulates NCAM (CD56): a novel mechanism for virus-induced tumor invasiveness
Abstract
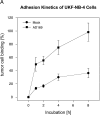
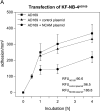
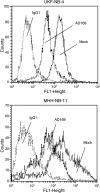
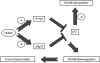
Pathologic data indicate that human cytomegalovirus (HCMV) infection might be associated with the pathogenesis of several human malignancies. However, no definitive evidence of a causal link between HCMV infection and cancer dissemination has been established to date. This study describes the modulation of the invasive behavior of NCAM-expressing tumor cell lines by HCMV. Neuroblastoma (NB) cells, persistently infected with the HCMV strain AD169 (UKF-NB-4AD169 and MHH-NB-11AD169), were added to endothelial cell monolayers and adhesion and penetration kinetics were measured. The 140- and 180-kDa isoforms of the adhesion receptor NCAM were evaluated by flow cytometry, Western blot, and reverse transcription-polymerase chain reaction (RT-PCR). The relevance of NCAM for tumor cell binding was proven by treating NB with NCAM antisense oligonucleotides or NCAM transfection. HCMV infection profoundly increased the number of adherent and penetrated NB, compared to controls. Surface expression of NCAM was significantly lower on UKF-NB-4AD169 and MHH-NB-11AD169, compared to mock-infected cells. Western-blot and RT-PCR demonstrated reduced protein and RNA levels of the 140- and 180-kDa isoform. An inverse correlation between NCAM expression and adhesion capacity of NB has been shown by antisense and transfection experiments. We conclude that HCMV infection leads to downregulation of NCAM receptors, which is associated with enhanced tumor cell invasiveness.
Copyright 2004 Neoplasia Press, Inc.
Figures








Similar articles
-
Chemoresistance induces enhanced adhesion and transendothelial penetration of neuroblastoma cells by down-regulating NCAM surface expression.BMC Cancer. 2006 Dec 21;6:294. doi: 10.1186/1471-2407-6-294. BMC Cancer. 2006. PMID: 17181871 Free PMC article.
-
[Cytomegalovirus infection as a possible factor in the progression of neuroblastoma].Klin Padiatr. 1999 Jul-Aug;211(4):310-3. doi: 10.1055/s-2008-1043806. Klin Padiatr. 1999. PMID: 10472568 German.
-
Persistent human cytomegalovirus infection induces drug resistance and alteration of programmed cell death in human neuroblastoma cells.Cancer Res. 1998 Jan 15;58(2):367-72. Cancer Res. 1998. PMID: 9443419
-
Oncomodulatory signals by regulatory proteins encoded by human cytomegalovirus: a novel role for viral infection in tumor progression.FEMS Microbiol Rev. 2004 Feb;28(1):59-77. doi: 10.1016/j.femsre.2003.07.005. FEMS Microbiol Rev. 2004. PMID: 14975530 Review.
-
The story of human cytomegalovirus and cancer: increasing evidence and open questions.Neoplasia. 2009 Jan;11(1):1-9. doi: 10.1593/neo.81178. Neoplasia. 2009. PMID: 19107226 Free PMC article. Review.
Cited by
-
The Human Cytomegalovirus, from Oncomodulation to Oncogenesis.Viruses. 2018 Aug 3;10(8):408. doi: 10.3390/v10080408. Viruses. 2018. PMID: 30081496 Free PMC article. Review.
-
Is HCMV a tumor promoter?Virus Res. 2011 May;157(2):193-203. doi: 10.1016/j.virusres.2010.10.026. Epub 2010 Oct 29. Virus Res. 2011. PMID: 21036194 Free PMC article. Review.
-
Extramedullary Multiple Myeloma: A Patient-Focused Review of the Pathogenesis of Bone Marrow Escape.World J Oncol. 2022 Oct;13(5):311-319. doi: 10.14740/wjon1521. Epub 2022 Oct 22. World J Oncol. 2022. PMID: 36406195 Free PMC article.
-
Is human cytomegalovirus a target in cancer therapy?Oncotarget. 2011 Dec;2(12):1329-38. doi: 10.18632/oncotarget.383. Oncotarget. 2011. PMID: 22246171 Free PMC article. Review.
-
Transcriptomic Changes of Bemisia tabaci Asia II 1 Induced by Chilli Leaf Curl Virus Trigger Infection and Circulation in Its Vector.Front Microbiol. 2022 Apr 28;13:890807. doi: 10.3389/fmicb.2022.890807. eCollection 2022. Front Microbiol. 2022. PMID: 35572639 Free PMC article.
References
-
- Cobbs CS, Harkins L, Samanta M, Gillespie GY, Bharara S, King PH, Nabors LB, Cobbs CG, Britt WJ. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 2002;62:3347–3350. - PubMed
-
- Samanta M, Harkins L, Klemm K, Britt WJ, Cobbs CS. High prevalence of human cytomegalovirus in prostatic intraepithelial neoplasia and prostatic carcinoma. J Urol. 2003;170:998–1002. - PubMed
-
- Harkins L, Volk AL, Samanta M, Mikolaenko I, Britt WJ, Bland KI, Cobbs CS. Specific localisation of human cytomegalovirus nucleic acids and proteins in human colorectal cancer. Lancet. 2002;360:1557–1563. - PubMed
-
- Cinatl J, Jr, Cinatl J, Vogel JU, Kotchetkov R, Driever PH, Kabickova H, Kornhuber B, Schwabe D, Doerr HW. Persistent human cytomegalovirus infection induces drug resistance and alteration of programmed cell death in human neuroblastoma cells. Cancer Res. 1998;58:367–372. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
