A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast
- PMID: 15256499
- PMCID: PMC478191
- DOI: 10.1101/gad.1178604
A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast
Abstract
Signaling molecules such as Cdc42 and mitogen-activated protein kinases (MAPKs) can function in multiple pathways in the same cell. Here, we propose one mechanism by which such factors may be directed to function in a particular pathway such that a specific response is elicited. Using genomic approaches, we identify a new component of the Cdc42- and MAPK-dependent signaling pathway that regulates filamentous growth (FG) in yeast. This factor, called Msb2, is a FG-pathway-specific factor that promotes differential activation of the MAPK for the FG pathway, Kss1. Msb2 is localized to polarized sites on the cell surface and interacts with Cdc42 and with the osmosensor for the high osmolarity glycerol response (HOG) pathway, Sho1. Msb2 is glycosylated and is a member of the mucin family, proteins that in mammalian cells promote disease resistance and contribute to metastasis in cancer cells. Remarkably, loss of the mucin domain of Msb2 causes hyperactivity of the FG pathway, demonstrating an inhibitory role for mucin domains in MAPK pathway activation. Taken together, our data suggest that Msb2 is a signaling mucin that interacts with general components, such as Cdc42 and Sho1, to promote their function in the FG pathway.
Copyright 2004 Cold Spring Harbor Laboratory Press ISSN
Figures
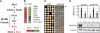
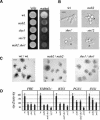





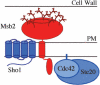
References
-
- Agrawal B., Gendler, S.J., and Longenecker, B.M. 1998a. The biological role of mucins in cellular interactions and immune regulation: Prospects for cancer immunotherapy. Mol. Med. Today 4: 397-403. - PubMed
-
- Agrawal B., Krantz, M.J., Parker, J., and Longenecker, B.M. 1998b. Expression of MUC1 mucin on activated human T cells: Implications for a role of MUC1 in normal immune regulation. Cancer Res. 58: 4079-4081. - PubMed
-
- Baldi P. and Long, A.D. 2001. A Bayesian framework for the analysis of microarray expression data: Regularized t-test and statistical inferences of gene changes. Bioinformatics 17: 509-519. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
