Clustering of DNA sequences in human promoters
- PMID: 15256515
- PMCID: PMC509265
- DOI: 10.1101/gr.1953904
Clustering of DNA sequences in human promoters
Abstract
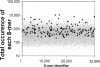
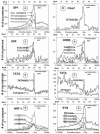
We have determined the distribution of each of the 65,536 DNA sequences that are eight bases long (8-mer) in a set of 13,010 human genomic promoter sequences aligned relative to the putative transcription start site (TSS). A limited number of 8-mers have peaks in their distribution (cluster), and most cluster within 100 bp of the TSS. The 156 DNA sequences exhibiting the greatest statistically significant clustering near the TSS can be placed into nine groups of related sequences. Each group is defined by a consensus sequence, and seven of these consensus sequences are known binding sites for the transcription factors (TFs) SP1, NF-Y, ETS, CREB, TBP, USF, and NRF-1. One sequence, which we named Clus1, is not a known TF binding site. The ninth sequence group is composed of the strand-specific Kozak sequence that clusters downstream of the TSS. An examination of the co-occurrence of these TF consensus sequences indicates a positive correlation for most of them except for sequences bound by TBP (the TATA box). Human mRNA expression data from 29 tissues indicate that the ETS, NRF-1, and Clus1 sequences that cluster are predominantly found in the promoters of housekeeping genes (e.g., ribosomal genes). In contrast, TATA is more abundant in the promoters of tissue-specific genes. This analysis identified eight DNA sequences in 5082 promoters that we suggest are important for regulating gene expression.
Copyright 2004 Cold Spring Harbor Laboratory Press ISSN
Figures







References
-
- Ashburner, M. and Lewis, S. 2002. On ontologies for biologists: The Gene Ontology: Untangling the web. Novartis Found. Symp. 247: 66–80. - PubMed
-
- Breathnach, R. and Chambon, P. 1981. Organization and expression of eucaryotic split genes coding for proteins. Annu. Rev. Biochem. 50: 349–383. - PubMed
-
- Brown, T.A. and McKnight, S.L. 1992. Specificities of protein–protein and protein–DNA interaction of GABP α and two newly defined ets-related proteins. Genes & Dev. 6: 2502–2512. - PubMed
WEB SITE REFERENCES
-
- http://genome.nci.nih.gov/publications/promoters; Supplemental data for this paper.
-
- http://transfac.gbf.de/TRANSFAC; the Transcription Factor Database.
-
- http://expression.gnf.org; GNF Gene Expression Atlas.
-
- http://genome.ucsc.edu/; UCSC Genome Bioinformatics site.
-
- http://dbtss.hgc.jp/index.html; database of TSS (DBTSS).
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
