Exploration of the conserved A+C wobble pair within the ribosomal peptidyl transferase center using affinity purified mutant ribosomes
- PMID: 15256541
- PMCID: PMC484164
- DOI: 10.1093/nar/gkh672
Exploration of the conserved A+C wobble pair within the ribosomal peptidyl transferase center using affinity purified mutant ribosomes
Abstract
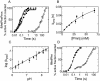
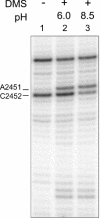
Protein synthesis in the ribosome's large subunit occurs within an active site comprised exclusively of RNA. Mutational studies of rRNA active site residues could provide valuable insight into the mechanism of peptide bond formation, but many of these mutations cause a dominant lethal phenotype, which prevents production of the homogeneous mutant ribosomes needed for analysis. We report a general method to affinity purify in vivo assembled 50S ribosomal subunits containing lethal active site mutations via a U1A protein-binding tag inserted onto the 23S rRNA. The expected pH-dependent formation of the A2450+C2063 wobble pair has made it a potential candidate for the pH-dependent conformational change that occurs within the ribosomal active site. Using this approach, the active site A2450+C2063 pair was mutated to the isosteric, but pH-independent, G2450*U2063 wobble pair, and 50S subunits containing the mutations were affinity purified. The G*U mutation caused the adjacent A2451 to become hyper-reactive to dimethylsulfate (DMS) modification in a pH-independent manner. Furthermore, the G*U mutation decreased both the rate of peptide bond formation and the affinity of the post-translocation complex for puromycin. The reaction rate (k(pep)) was reduced approximately 200-fold for both puromycin and the natural aminoacyl-tRNA A-site substrate. The mutations also substantially altered the pH dependence of the reaction. Mutation of this base pair has significant deleterious effects upon peptidyl transferase activity, but because G*U mutation disrupts several tertiary contacts with the wobble pair, the assignment of A2450 as the active site residue with the neutral pK(a) important for the peptidyl transferase reaction cannot be fully supported or excluded based upon these data.
Figures



Similar articles
-
The role of the universally conserved A2450-C2063 base pair in the ribosomal peptidyl transferase center.Nucleic Acids Res. 2010 Aug;38(14):4844-55. doi: 10.1093/nar/gkq213. Epub 2010 Apr 7. Nucleic Acids Res. 2010. PMID: 20375101 Free PMC article.
-
The A2453-C2499 wobble base pair in Escherichia coli 23S ribosomal RNA is responsible for pH sensitivity of the peptidyltransferase active site conformation.Nucleic Acids Res. 2004 Oct 12;32(18):5512-8. doi: 10.1093/nar/gkh888. Print 2004. Nucleic Acids Res. 2004. PMID: 15479786 Free PMC article.
-
Ribosomal peptidyl transferase can withstand mutations at the putative catalytic nucleotide.Nature. 2001 May 24;411(6836):498-501. doi: 10.1038/35078113. Nature. 2001. PMID: 11373685
-
Importance of tRNA interactions with 23S rRNA for peptide bond formation on the ribosome: studies with substrate analogs.Biol Chem. 2007 Jul;388(7):687-91. doi: 10.1515/BC.2007.077. Biol Chem. 2007. PMID: 17570820 Review.
-
Peptide bond formation on the ribosome: structure and mechanism.Curr Opin Struct Biol. 2003 Jun;13(3):334-40. doi: 10.1016/s0959-440x(03)00065-4. Curr Opin Struct Biol. 2003. PMID: 12831884 Review.
Cited by
-
The role of the universally conserved A2450-C2063 base pair in the ribosomal peptidyl transferase center.Nucleic Acids Res. 2010 Aug;38(14):4844-55. doi: 10.1093/nar/gkq213. Epub 2010 Apr 7. Nucleic Acids Res. 2010. PMID: 20375101 Free PMC article.
-
Generation of chemically engineered ribosomes for atomic mutagenesis studies on protein biosynthesis.Nat Protoc. 2011 May;6(5):580-92. doi: 10.1038/nprot.2011.306. Epub 2011 Apr 7. Nat Protoc. 2011. PMID: 21527916
-
A single-step method for purification of active His-tagged ribosomes from a genetically engineered Escherichia coli.Nucleic Acids Res. 2009 Feb;37(2):e15. doi: 10.1093/nar/gkn992. Epub 2008 Dec 11. Nucleic Acids Res. 2009. PMID: 19074194 Free PMC article.
-
Oxidative stress damages rRNA inside the ribosome and differentially affects the catalytic center.Nucleic Acids Res. 2018 Feb 28;46(4):1945-1957. doi: 10.1093/nar/gkx1308. Nucleic Acids Res. 2018. PMID: 29309687 Free PMC article.
-
Towards Accurate Prediction of Protonation Equilibrium of Nucleic Acids.J Phys Chem Lett. 2013 Mar 7;4(5):760-766. doi: 10.1021/jz400078d. Epub 2013 Feb 12. J Phys Chem Lett. 2013. PMID: 23526474 Free PMC article.
References
-
- Nissen P., Hansen,J., Ban,N., Moore,P.B. and Steitz,T.A. (2000) The structural basis of ribosome activity in peptide bond synthesis. Science, 289, 920–930. - PubMed
-
- Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. - PubMed
-
- Green R. and Noller,H.F. (1997) Ribosomes and translation. Annu. Rev. Biochem., 66, 679–716. - PubMed
-
- Yusupov M.M., Yusupova,G.Z., Baucom,A., Lieberman,K., Earnest,T.N., Cate,J.H. and Noller,H.F. (2001) Crystal structure of the ribosome at 5.5 Å resolution. Science, 292, 883–896. - PubMed
-
- Harms J., Schluenzen,F., Zarivach,R., Bashan,A., Gat,S., Agmon,I., Bartels,H., Franceschi,F. and Yonath,A. (2001) High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell, 107, 679–688. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

