Variable var transition rates underlie antigenic variation in malaria
- PMID: 15256597
- PMCID: PMC503751
- DOI: 10.1073/pnas.0402347101
Variable var transition rates underlie antigenic variation in malaria
Abstract
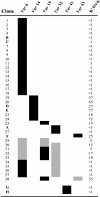
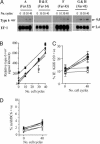
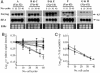
Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) is expressed on the surface of infected erythrocytes where it plays a central role in both infected erythrocytes cytoadhesion and immune evasion. Switches in clonal expression of PfEMP1 result in antigenic variation that facilitates long-term chronic infection of the host. The var gene family encodes PfEMP1 variants, with transcriptional switching between different var variants providing the molecular basis for antigenic variation. Despite the importance of var transcriptional switching in the evasion of the immune response, little is known about the way in which this process is regulated. Here we report the measurement of transition on and off rates for a series of var gene variants. We find (i) that on and off rates for a given variant are dissimilar, (ii) that these rates vary dramatically among different variants, and (iii) that in isogenic clones expressing the same var gene, both on and off rates are constant and appear to be an intrinsic property of that particular gene. These data would suggest that the information that determines the probability of the activation or silencing of var genes is present in their surrounding DNA. Furthermore, some transitions appear to be disallowed depending on the recent variant antigen expression history of the parasite clone. These findings have important implications for both the underlying molecular mechanisms of antigenic variation and the processes that promote chronicity of infection in vivo.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

