Nucleosomes containing the histone variant H2A.Bbd organize only 118 base pairs of DNA
- PMID: 15257289
- PMCID: PMC514500
- DOI: 10.1038/sj.emboj.7600316
Nucleosomes containing the histone variant H2A.Bbd organize only 118 base pairs of DNA
Abstract
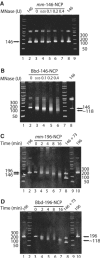
H2A.Bbd is an unusual histone variant whose sequence is only 48% conserved compared to major H2A. The major sequence differences are in the docking domain that tethers the H2A-H2B dimer to the (H3-H4)(2) tetramer; in addition, the C-terminal tail is absent in H2A.Bbd. We assembled nucleosomes in which H2A is replaced by H2A.Bbd (Bbd-NCP), and found that Bbd-NCP had a more relaxed structure in which only 118+/-2 bp of DNA is protected against digestion with micrococcal nuclease. The absence of fluorescence resonance energy transfer between the ends of the DNA in Bbd-NCP indicates that the distance between the DNA ends is increased significantly. The Bbd docking domain is largely responsible for this behavior, as shown by domain-swap experiments. Bbd-containing nucleosomal arrays repress transcription from a natural promoter, and this repression can be alleviated by transcriptional activators Tax and CREB. The structural properties of Bbd-NCP described here have important implications for the in vivo function of this histone variant and are consistent with its proposed role in transcriptionally active chromatin.
Figures




References
-
- Akey CW, Luger K (2003) Histone chaperones and nucleosome assembly. Curr Opin Struct Biol 13: 6–14 - PubMed
-
- Becker PB, Horz W (2002) ATP-dependent nucleosome remodeling. Annu Rev Biochem 71: 247–273 - PubMed
-
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D (2003) FACT facilitates transcription-dependent nucleosome alteration. Science 301: 1090–1093 - PubMed
-
- Brooks W, Jackson V (1994) The rapid transfer and selective association of histones H2A and H2B onto negatively coiled DNA at physiological ionic strength. J Biol Chem 269: 18155–18166 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

