High frequency of mosaicism among patients with neurofibromatosis type 1 (NF1) with microdeletions caused by somatic recombination of the JJAZ1 gene
- PMID: 15257518
- PMCID: PMC1182020
- DOI: 10.1086/423624
High frequency of mosaicism among patients with neurofibromatosis type 1 (NF1) with microdeletions caused by somatic recombination of the JJAZ1 gene
Abstract
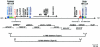
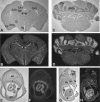
Detailed analyses of 20 patients with sporadic neurofibromatosis type 1 (NF1) microdeletions revealed an unexpected high frequency of somatic mosaicism (8/20 [40%]). This proportion of mosaic deletions is much higher than previously anticipated. Of these deletions, 16 were identified by a screen of unselected patients with NF1. None of the eight patients with mosaic deletions exhibited the mental retardation and facial dysmorphism usually associated with NF1 microdeletions. Our study demonstrates the importance of a general screening for NF1 deletions, regardless of a special phenotype, because of a high estimated number of otherwise undetected mosaic NF1 microdeletions. In patients with mosaicism, the proportion of cells with the deletion was 91%-100% in peripheral leukocytes but was much lower (51%-80%) in buccal smears or peripheral skin fibroblasts. Therefore, the analysis of other tissues than blood is recommended, to exclude mosaicism with normal cells in patients with NF1 microdeletions. Furthermore, our study reveals breakpoint heterogeneity. The classic 1.4-Mb deletion was found in 13 patients. These type I deletions encompass 14 genes and have breakpoints in the NF1 low-copy repeats. However, we identified a second major type of NF1 microdeletion, which spans 1.2 Mb and affects 13 genes. This type II deletion was found in 8 (38%) of 21 patients and is mediated by recombination between the JJAZ1 gene and its pseudogene. The JJAZ1 gene, which is completely deleted in patients with type I NF1 microdeletions and is disrupted in deletions of type II, is highly expressed in brain structures associated with learning and memory. Thus, its haploinsufficiency might contribute to mental impairment in patients with constitutional NF1 microdeletions. Conspicuously, seven of the eight mosaic deletions are of type II, whereas only one was a classic type I deletion. Therefore, the JJAZ1 gene is a preferred target of strand exchange during mitotic nonallelic homologous recombination. Although type I NF1 microdeletions occur by interchromosomal recombination during meiosis, our findings imply that type II deletions are mediated by intrachromosomal recombination during mitosis. Thus, NF1 microdeletions acquired during mitotic cell divisions differ from those occurring in meiosis and are caused by different mechanisms.
Figures
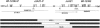


References
Electronic-Database Information
-
- BACPAC Resource Center Home Page, http://bacpac.chori.org/
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for PAC 2349P21 [accession number AC127024], BAC 307A16 [accession number AC003041], BAC 55A13 [accession number AC015651], cDNA clone 6513835 [accession number BQ964374], BAC 640N20 [accession number AC090616], RP11-271K11 [accession number AC005562], CTD-2349P21 [accession number AC127024], RP11-785C15 [accession number AC109516], RP11-142O6 [accession number AC079915], RP11-805L22 [accession number AC007923], HCIT-307A16 [accession number AC003041], and RP11-55J8 [accession number AC015941])
-
- Karolinska Institutet Center for Genomics Research, http://www.cgr.ki.se/cgr/groups/sonnhammer/Dotter.html (for Dotter program)
-
- National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for NF1) - PubMed
References
-
- Chance PF, Abbas N, Lensch MW, Pentao L, Roa BB, Patel PI, Lupski JR (1994) Two autosomal dominant neuropathies result from reciprocal DNA duplication/deletion of a region on chromosome 17. Hum Mol Genet 3:223–228 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

