The homogentisate pathway: a central catabolic pathway involved in the degradation of L-phenylalanine, L-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida
- PMID: 15262943
- PMCID: PMC451635
- DOI: 10.1128/JB.186.15.5062-5077.2004
The homogentisate pathway: a central catabolic pathway involved in the degradation of L-phenylalanine, L-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida
Abstract
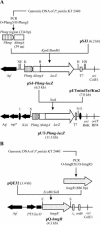
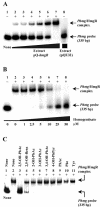
Pseudomonas putida metabolizes Phe and Tyr through a peripheral pathway involving hydroxylation of Phe to Tyr (PhhAB), conversion of Tyr into 4-hydroxyphenylpyruvate (TyrB), and formation of homogentisate (Hpd) as the central intermediate. Homogentisate is then catabolized by a central catabolic pathway that involves three enzymes, homogentisate dioxygenase (HmgA), fumarylacetoacetate hydrolase (HmgB), and maleylacetoacetate isomerase (HmgC), finally yielding fumarate and acetoacetate. Whereas the phh, tyr, and hpd genes are not linked in the P. putida genome, the hmgABC genes appear to form a single transcriptional unit. Gel retardation assays and lacZ translational fusion experiments have shown that hmgR encodes a specific repressor that controls the inducible expression of the divergently transcribed hmgABC catabolic genes, and homogentisate is the inducer molecule. Footprinting analysis revealed that HmgR protects a region in the Phmg promoter that spans a 17-bp palindromic motif and an external direct repetition from position -16 to position 29 with respect to the transcription start site. The HmgR protein is thus the first IclR-type regulator that acts as a repressor of an aromatic catabolic pathway. We engineered a broad-host-range mobilizable catabolic cassette harboring the hmgABC, hpd, and tyrB genes that allows heterologous bacteria to use Tyr as a unique carbon and energy source. Remarkably, we show here that the catabolism of 3-hydroxyphenylacetate in P. putida U funnels also into the homogentisate central pathway, revealing that the hmg cluster is a key catabolic trait for biodegradation of a small number of aromatic compounds.
Figures







References
-
- Adachi, K., Y. Iwayama, H. Tanioka, and Y. Takeda. 1966. Purification and properties of homogentisate oxygenase from Pseudomonas fluorescens. Biochim. Biophys. Acta 118:88-97. - PubMed
-
- Bayley, S. A., C. J. Duggleby, M. J. Worsey, P. A. Williams, K. G. Hardy, and P. Broda. 1977. Two modes of loss of the Tol function from Pseudomonas putida mt-2. Mol. Gen. Genet. 154:203-204. - PubMed
-
- Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementary analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
-
- Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, N. Zafar, L. Zhou, J. Liu, Q. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Van Aken, T. V. Feldblyum, M. D'Ascenzo, W. L. Deng, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martín, D. J. Schneider, X. Tang, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100:10181-10186. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

