Characterization of Arabidopsis glutamine phosphoribosyl pyrophosphate amidotransferase-deficient mutants
- PMID: 15266056
- PMCID: PMC519050
- DOI: 10.1104/pp.104.040956
Characterization of Arabidopsis glutamine phosphoribosyl pyrophosphate amidotransferase-deficient mutants
Abstract
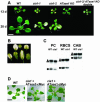
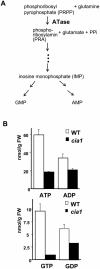
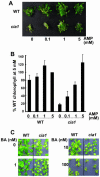
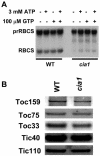
Using a transgene-based screening, we previously isolated several Arabidopsis mutants defective in protein import into chloroplasts. Positional cloning of one of the loci, CIA1, revealed that CIA1 encodes Gln phosphoribosyl pyrophosphate amidotransferase 2 (ATase2), one of the three ATase isozymes responsible for the first committed step of de novo purine biosynthesis. The cia1 mutant had normal green cotyledons but small and albino/pale-green mosaic leaves. Adding AMP, but not cytokinin or NADH, to plant liquid cultures partially complemented the mutant phenotypes. Both ATase1 and ATase2 were localized to chloroplasts. Overexpression of ATase1 fully complemented the ATase2-deficient phenotypes. A T-DNA insertion knockout mutant of the ATase1 gene was also obtained. The mutant was indistinguishable from the wild type. A double mutant of cia1/ATase1-knockout had the same phenotype as cia1, suggesting at least partial gene redundancy between ATase1 and ATase2. Characterizations of the cia1 mutant revealed that mutant leaves had slightly smaller cell size but only half the cell number of wild-type leaves. This phenotype confirms the role of de novo purine biosynthesis in cell division. Chloroplasts isolated from the cia1 mutant imported proteins at an efficiency less than 50% that of wild-type chloroplasts. Adding ATP and GTP to isolated mutant chloroplasts could not restore the import efficiency. We conclude that de novo purine biosynthesis is not only important for cell division, but also for chloroplast biogenesis.
Figures




Similar articles
-
The leaf reticulate mutant dov1 is impaired in the first step of purine metabolism.Mol Plant. 2012 Nov;5(6):1227-41. doi: 10.1093/mp/sss045. Epub 2012 Apr 24. Mol Plant. 2012. PMID: 22532604
-
Purine biosynthetic enzyme ATase2 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis.Photosynth Res. 2015 Dec;126(2-3):285-300. doi: 10.1007/s11120-015-0131-z. Epub 2015 Apr 3. Photosynth Res. 2015. PMID: 25837856
-
Molecular analysis of "de novo" purine biosynthesis in solanaceous species and in Arabidopsis thaliana.Front Biosci. 2004 May 1;9:1803-16. doi: 10.2741/1344. Front Biosci. 2004. PMID: 14977588
-
Feedback inhibition of amidophosphoribosyltransferase regulates the rate of cell growth via purine nucleotide, DNA, and protein syntheses.J Biol Chem. 2001 Jun 15;276(24):21285-91. doi: 10.1074/jbc.M011103200. Epub 2001 Apr 4. J Biol Chem. 2001. PMID: 11290738
-
The novel protein DELAYED PALE-GREENING1 is required for early chloroplast biogenesis in Arabidopsis thaliana.Sci Rep. 2016 May 10;6:25742. doi: 10.1038/srep25742. Sci Rep. 2016. PMID: 27160321 Free PMC article.
Cited by
-
Proteomic Analysis of the Function of a Novel Cold-Regulated Multispanning Transmembrane Protein COR413-PM1 in Arabidopsis.Int J Mol Sci. 2018 Aug 29;19(9):2572. doi: 10.3390/ijms19092572. Int J Mol Sci. 2018. PMID: 30158496 Free PMC article.
-
Chemical genetic identification of glutamine phosphoribosylpyrophosphate amidotransferase as the target for a novel bleaching herbicide in Arabidopsis.Plant Physiol. 2007 Jul;144(3):1292-304. doi: 10.1104/pp.107.099705. Epub 2007 Jun 1. Plant Physiol. 2007. PMID: 17616508 Free PMC article.
-
Phosphoribosyltransferases and Their Roles in Plant Development and Abiotic Stress Response.Int J Mol Sci. 2023 Jul 23;24(14):11828. doi: 10.3390/ijms241411828. Int J Mol Sci. 2023. PMID: 37511586 Free PMC article. Review.
-
Fine mapping and molecular characterization of the virescent gene vsp in Upland cotton (Gossypium hirsutum).Theor Appl Genet. 2019 Jul;132(7):2069-2086. doi: 10.1007/s00122-019-03338-9. Epub 2019 Apr 5. Theor Appl Genet. 2019. PMID: 30953093
-
Crystal Structure of the Chloroplastic Glutamine Phosphoribosylpyrophosphate Amidotransferase GPRAT2 From Arabidopsis thaliana.Front Plant Sci. 2020 Feb 27;11:157. doi: 10.3389/fpls.2020.00157. eCollection 2020. Front Plant Sci. 2020. PMID: 32174940 Free PMC article.
References
-
- Ashihara H (1983) Changes in activities of purine salvage and ureide synthesis during germination of black gram (Phaseolus mungo) seeds. Z Pflanzenphysiol 113: 47–60
-
- Boland M, Schubert K (1983) Biosynthesis of purines by a proplastid fraction from soybean nodules. Arch Biochem Biophys 220: 179–187 - PubMed
-
- Boldt R, Zrenner R (2003) Purine and pyrimidine biosynthesis in higher plants. Physiol Plant 117: 297–304 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

