Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver
- PMID: 15266058
- PMCID: PMC509189
- DOI: 10.1073/pnas.0404297101
Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver
Abstract
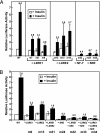
Transcription of the gene encoding sterol regulatory element-binding protein 1c (SREBP-1c) is known to be activated by insulin in the liver. The resultant SREBP-1c protein activates transcription of the genes required for fatty acid synthesis. Here, we use SREBP-1c promoter reporter constructs to dissect the mechanism of insulin activation in freshly isolated rat hepatocytes. The data show that a complete insulin response (increase of 6- to 11-fold) requires two binding sites for liver X receptors (LXRs), which are nuclear receptors that are activated by oxygenated sterols. Disruption of these binding sites did not lower basal transcription but severely reduced the response to insulin. In contrast, disruption of the closely linked binding sites for SREBPs and nuclear factor Y lowered basal transcription drastically but still permitted a 4- to 7-fold increase in response to insulin. Arachidonic acid, an inhibitor of LXR activation, blocked the response to insulin. We conclude that insulin activates the SREBP-1c promoter primarily by increasing the activity of LXRs, possibly through production of a ligand that activates LXRs or their heterodimerizing partner, the retinoid X receptor. Nuclear SREBPs and nuclear factor Y play permissive roles.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

