Hydrogen photoproduction is attenuated by disruption of an isoamylase gene in Chlamydomonas reinhardtii
- PMID: 15269330
- PMCID: PMC519204
- DOI: 10.1105/tpc.104.021972
Hydrogen photoproduction is attenuated by disruption of an isoamylase gene in Chlamydomonas reinhardtii
Abstract
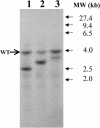
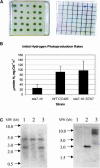
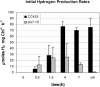
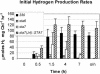
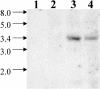
DNA insertional transformants of Chlamydomonas reinhardtii were screened chemochromically for attenuated H(2) production. One mutant, displaying low H(2) gas photoproduction, has a nonfunctional copy of a gene that shows high homology to the family of isoamylase genes found in several photosynthetic organisms. DNA gel blotting and gene complementation were used to link this isoamylase gene to previously characterized nontagged sta7 mutants. This mutant is therefore denoted sta7-10. In C. reinhardtii, the STA7 isoamylase gene is important for the accumulation of crystalline starch, and the sta7-10 mutant reported here contains <3% of the glucose found in insoluble starch when compared with wild-type control cells. Hydrogen photoproduction rates, induced after several hours of dark, anaerobic treatment, are attenuated in sta7 mutants. RNA gel blot analysis indicates that the mRNA transcripts for both the HydA1 and HydA2 [Fe]-hydrogenase genes are expressed in the sta7-10 mutant at greater than wild-type levels 0.5 h after anaerobic induction. However, after 1.5 h, transcript levels of both HydA1 and HydA2 begin to decline rapidly and reach nearly undetectable levels after 7 h. In wild-type cells, the hydrogenase transcripts accumulate more slowly, reach a plateau after 4 h of anaerobic treatment, and maintain the same level of expression for >7 h under anaerobic incubation. Complementation of mutant cells with genomic DNA corresponding to the STA7 gene restores both the starch accumulation and H(2) production phenotypes. The results indicate that STA7 and starch metabolism play an important role in C. reinhardtii H(2) photoproduction. Moreover, the results indicate that mere anaerobiosis is not sufficient to maintain hydrogenase gene expression without the underlying physiology, an important aspect of which is starch metabolism.
Figures





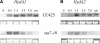

Similar articles
-
Increased lipid accumulation in the Chlamydomonas reinhardtii sta7-10 starchless isoamylase mutant and increased carbohydrate synthesis in complemented strains.Eukaryot Cell. 2010 Aug;9(8):1251-61. doi: 10.1128/EC.00075-10. Epub 2010 Jun 18. Eukaryot Cell. 2010. PMID: 20562225 Free PMC article.
-
Identification of genes required for hydrogenase activity in Chlamydomonas reinhardtii.Biochem Soc Trans. 2005 Feb;33(Pt 1):102-4. doi: 10.1042/BST0330102. Biochem Soc Trans. 2005. PMID: 15667277
-
Genetic disruption of both Chlamydomonas reinhardtii [FeFe]-hydrogenases: Insight into the role of HYDA2 in H₂ production.Biochem Biophys Res Commun. 2012 Jan 13;417(2):704-9. doi: 10.1016/j.bbrc.2011.12.002. Epub 2011 Dec 8. Biochem Biophys Res Commun. 2012. PMID: 22177948
-
Photosynthetic H2 metabolism in Chlamydomonas reinhardtii (unicellular green algae).Planta. 2007 Oct;226(5):1075-86. doi: 10.1007/s00425-007-0609-9. Epub 2007 Aug 25. Planta. 2007. PMID: 17721788 Review.
-
Approaches to developing biological H(2)-photoproducing organisms and processes.Biochem Soc Trans. 2005 Feb;33(Pt 1):70-2. doi: 10.1042/BST0330070. Biochem Soc Trans. 2005. PMID: 15667268 Review.
Cited by
-
Plastidial Expression of Type II NAD(P)H Dehydrogenase Increases the Reducing State of Plastoquinones and Hydrogen Photoproduction Rate by the Indirect Pathway in Chlamydomonas reinhardtii1.Plant Physiol. 2014 Jul;165(3):1344-1352. doi: 10.1104/pp.114.240432. Epub 2014 May 12. Plant Physiol. 2014. PMID: 24820024 Free PMC article.
-
Molecular characterization of an isoamylase 1-type starch debranching enzyme (DBEI) in grain amaranth (Amaranthus cruentus L.).Mol Biol Rep. 2014 Dec;41(12):7857-64. doi: 10.1007/s11033-014-3679-3. Epub 2014 Aug 29. Mol Biol Rep. 2014. PMID: 25167854
-
Characterization of Function of the GlgA2 Glycogen/Starch Synthase in Cyanobacterium sp. Clg1 Highlights Convergent Evolution of Glycogen Metabolism into Starch Granule Aggregation.Plant Physiol. 2016 Jul;171(3):1879-92. doi: 10.1104/pp.16.00049. Epub 2016 May 19. Plant Physiol. 2016. PMID: 27208262 Free PMC article.
-
Increased lipid accumulation in the Chlamydomonas reinhardtii sta7-10 starchless isoamylase mutant and increased carbohydrate synthesis in complemented strains.Eukaryot Cell. 2010 Aug;9(8):1251-61. doi: 10.1128/EC.00075-10. Epub 2010 Jun 18. Eukaryot Cell. 2010. PMID: 20562225 Free PMC article.
-
Rewiring hydrogenase-dependent redox circuits in cyanobacteria.Proc Natl Acad Sci U S A. 2011 Mar 8;108(10):3941-6. doi: 10.1073/pnas.1016026108. Epub 2011 Feb 22. Proc Natl Acad Sci U S A. 2011. PMID: 21368150 Free PMC article.
References
-
- Adam, M., and Loppes, R. (1998). Use of the ARG7 gene as an insertional mutagen to clone PHON24, a gene required for derepressible neutral phosphatase activity in Chlamydomonas reinhardtii. Mol. Gen. Genet. 258, 123–132. - PubMed
-
- Adams, M.W.W. (1990). The structure and mechanism of iron-hydrogenases. Biochim. Biophys. Acta 1020, 115–145. - PubMed
-
- Asada, Y., and Miyake, J. (1999). Photobiological hydrogen production. J. Biosci. Bioeng. 88, 1–6. - PubMed
-
- Ball, S. (1998). Regulation of starch biosynthesis. In The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas, Vol. 7, J.D. Rochaix, M. Goldschmiidt-Clermont, and S. Merchant, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 549–567.
-
- Ball, S., Guan, H.P., James, M., Myers, A., Keeling, P., Mouille, G., Buleon, A., Colonna, P., and Preiss, J. (1996). From glycogen to amylopectin: A model for the biogenesis of the plant starch granule. Cell 86, 349–352. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

