Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis
- PMID: 15269332
- PMCID: PMC519200
- DOI: 10.1105/tpc.104.022236
Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis
Abstract
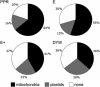
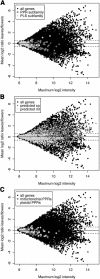
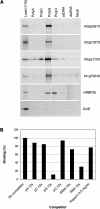
The complete sequence of the Arabidopsis thaliana genome revealed thousands of previously unsuspected genes, many of which cannot be ascribed even putative functions. One of the largest and most enigmatic gene families discovered in this way is characterized by tandem arrays of pentatricopeptide repeats (PPRs). We describe a detailed bioinformatic analysis of 441 members of the Arabidopsis PPR family plus genomic and genetic data on the expression (microarray data), localization (green fluorescent protein and red fluorescent protein fusions), and general function (insertion mutants and RNA binding assays) of many family members. The basic picture that arises from these studies is that PPR proteins play constitutive, often essential roles in mitochondria and chloroplasts, probably via binding to organellar transcripts. These results confirm, but massively extend, the very sparse observations previously obtained from detailed characterization of individual mutants in other organisms.
Figures
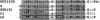




References
-
- Akashi, K., Grandjean, O., and Small, I. (1998). Potential dual targeting of an Arabidopsis archaebacterial-like histidyl-tRNA synthetase to mitochondria and chloroplasts. FEBS Lett. 431, 39–44. - PubMed
-
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. - PubMed
-
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
-
- Aubourg, S., Boudet, N., Kreis, M., and Lecharny, A. (2000). In Arabidopsis thaliana, 1% of the genome codes for a novel protein family unique to plants. Plant Mol. Biol. 42, 603–613. - PubMed
-
- Barkan, A., and Goldschmidt-Clermont, M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82, 559–572. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

