Distinct classes of glyoxalase I: metal specificity of the Yersinia pestis, Pseudomonas aeruginosa and Neisseria meningitidis enzymes
- PMID: 15270717
- PMCID: PMC1134094
- DOI: 10.1042/BJ20041006
Distinct classes of glyoxalase I: metal specificity of the Yersinia pestis, Pseudomonas aeruginosa and Neisseria meningitidis enzymes
Abstract
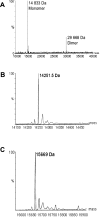
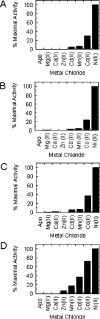
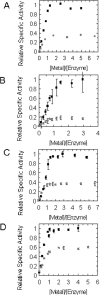
The metalloisomerase glyoxalase I (GlxI) catalyses the conversion of methylglyoxal-glutathione hemithioacetal and related derivatives into the corresponding thioesters. In contrast with the previously characterized GlxI enzymes of Homo sapiens, Pseudomonas putida and Saccharomyces cerevisiae, we recently determined that Escherichia coli GlxI surprisingly did not display Zn2+-activation, but instead exhibited maximal activity with Ni2+. To investigate whether non-Zn2+ activation defines a distinct, previously undocumented class of GlxI enzymes, or whether the E. coli GlxI is an exception to the previously established Zn2+-activated GlxI, we have cloned and characterized the bacterial GlxI from Yersinia pestis, Pseudomonas aeruginosa and Neisseria meningitidis. The metal-activation profiles for these additional GlxIs firmly establish the existence of a non-Zn2+-dependent grouping within the general category of GlxI enzymes. This second, established class of metal activation was formerly unidentified for this metalloenzyme. Amino acid sequence comparisons indicate a more extended peptide chain in the Zn2+-dependent forms of GlxI (H. sapiens, P. putida and S. cerevisiae), compared with the GlxI enzymes of E. coli, Y. pestis, P. aeruginosa and N. meningitidis. The longer sequence is due in part to the presence of additional regions situated fairly close to the metal ligands in the Zn2+-dependent forms of the lyase. With respect to sequence alignments, these inserts may potentially contribute to defining the metal specificity of GlxI at a structural level.
Figures

Similar articles
-
Pseudomonas aeruginosa contains multiple glyoxalase I-encoding genes from both metal activation classes.Biochim Biophys Acta. 2007 Jun;1774(6):756-63. doi: 10.1016/j.bbapap.2007.04.005. Epub 2007 Apr 20. Biochim Biophys Acta. 2007. PMID: 17513180
-
Overproduction and characterization of a dimeric non-zinc glyoxalase I from Escherichia coli: evidence for optimal activation by nickel ions.Biochemistry. 1998 Jun 16;37(24):8754-63. doi: 10.1021/bi972791w. Biochemistry. 1998. PMID: 9628737
-
Escherichia coli glyoxalase II is a binuclear zinc-dependent metalloenzyme.Arch Biochem Biophys. 2007 Mar 1;459(1):20-6. doi: 10.1016/j.abb.2006.11.024. Epub 2006 Dec 6. Arch Biochem Biophys. 2007. PMID: 17196158
-
Microbial glyoxalase enzymes: metalloenzymes controlling cellular levels of methylglyoxal.Drug Metabol Drug Interact. 2008;23(1-2):29-50. doi: 10.1515/dmdi.2008.23.1-2.29. Drug Metabol Drug Interact. 2008. PMID: 18533363 Review.
-
Glyoxalase I inhibitors in cancer chemotherapy.Biochem Soc Trans. 2003 Dec;31(Pt 6):1378-82. doi: 10.1042/bst0311378. Biochem Soc Trans. 2003. PMID: 14641067 Review.
Cited by
-
Arabidopsis thaliana Contains Both Ni2+ and Zn2+ Dependent Glyoxalase I Enzymes and Ectopic Expression of the Latter Contributes More towards Abiotic Stress Tolerance in E. coli.PLoS One. 2016 Jul 14;11(7):e0159348. doi: 10.1371/journal.pone.0159348. eCollection 2016. PLoS One. 2016. PMID: 27415831 Free PMC article.
-
Methylglyoxal metabolism in trypanosomes and leishmania.Semin Cell Dev Biol. 2011 May;22(3):271-7. doi: 10.1016/j.semcdb.2011.02.001. Epub 2011 Feb 15. Semin Cell Dev Biol. 2011. PMID: 21310261 Free PMC article. Review.
-
Structure of the novel monomeric glyoxalase I from Zea mays.Acta Crystallogr D Biol Crystallogr. 2015 Oct;71(Pt 10):2009-20. doi: 10.1107/S1399004715015205. Epub 2015 Sep 26. Acta Crystallogr D Biol Crystallogr. 2015. PMID: 26457425 Free PMC article.
-
Bioinformatics of Metalloproteins and Metalloproteomes.Molecules. 2020 Jul 24;25(15):3366. doi: 10.3390/molecules25153366. Molecules. 2020. PMID: 32722260 Free PMC article. Review.
-
Trypanothione-dependent glyoxalase I in Trypanosoma cruzi.Biochem J. 2006 Dec 1;400(2):217-23. doi: 10.1042/BJ20060882. Biochem J. 2006. PMID: 16958620 Free PMC article.
References
-
- Neuberg C. The destruction of lactic aldehyde and methylglyoxal by animal organisms. Biochem. Z. 1931;49:502–506.
-
- Dakin H. D., Dudley H. W. An enzyme concerned with the formation of hydroxy acids from ketonic aldehydes. J. Biol. Chem. 1913;14:155–157.
-
- Thornalley P. J. Glyoxalase I – structure, function and a critical role in the enzymatic defence against glycation. Biochem. Soc. Trans. 2003;31:1343–1348. - PubMed
-
- Becker K., Rahlfs S., Nickel C., Schirmer R. H. Glutathione – functions and metabolism in the malarial parasite Plasmodium falciparum. Biol. Chem. 2003;384:551–566. - PubMed
-
- Rose I. A., Nowick J. S. Methylglyoxal synthetase, enol-pyruvaldehyde, glutathione and the glyoxalase system. J. Am. Chem. Soc. 2002;124:13047–13052. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

