Interaction of Chlamydia trachomatis serovar L2 with the host autophagic pathway
- PMID: 15271937
- PMCID: PMC470602
- DOI: 10.1128/IAI.72.8.4751-4762.2004
Interaction of Chlamydia trachomatis serovar L2 with the host autophagic pathway
Abstract
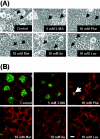
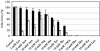
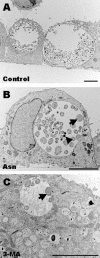
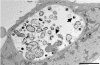
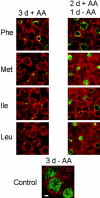
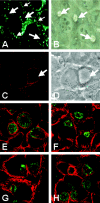
Chlamydiae are obligate intracellular pathogens that replicate within a membrane-bound compartment (the inclusion) and are associated with important human diseases, such as trachoma, pneumonia, and atherosclerosis. We have examined the interaction of the host autophagic pathway with Chlamydia trachomatis serovar L2 by using the specific autophagosomal stain monodansylcadaverine, antibodies to autophagosome-associated markers, and traditionally used autophagic inhibitors, particularly 3-methyladenine and amino acids. Chlamydial inclusions did not sequester monodansylcadaverine, suggesting absence of fusion with autophagosomes. Interestingly, exposure of cultures infected for 19 h to 3-methyladenine or single amino acids until the end of infection (44 h) caused various degrees of abnormalities in the inclusion maturation and in the progeny infectivity. Incubation of host cells with chemicals throughout the entire period of infection modulated the growth of Chlamydia even more dramatically. Remarkably, autophagosomal markers MAP-LC3 and calreticulin were redistributed to the inclusion of Chlamydia, a process that appears to be sensitive to 3-methyladenine and some amino acids. The present data indicate the lack of autophagosomal fusion with the inclusion because it was devoid of monodansylcadaverine and no distinct rim of autophagosomal protein-specific staining around the inclusion could be observed. However, high sensitivity of Chlamydia to conditions that could inhibit host autophagic pathway and the close association of MAP-LC3 and calreticulin with the inclusion membrane still suggest a potential role of host autophagy in the pathogenesis of Chlamydia.
Figures

References
-
- Allan, I., and J. H. Pearce. 1983. Differential amino acid utilization by Chlamydia psittaci (strain guinea pig inclusion conjunctivitis) and its regulatory effect on chlamydial growth. J. Gen. Microbiol. 129:1991-2000. - PubMed
-
- Allan, I., T. P. Hatch, and J. H. Pearce. 1985. Influence of cysteine deprivation on chlamydial differentiation from reproductive to infective life-cycle forms. J. Gen. Microbiol. 131:3171-3177. - PubMed
-
- Al-Younes, H. M., T. Rudel, and T. F. Meyer. 1999. Characterization and intracellular trafficking pattern of vacuoles containing Chlamydia pneumoniae in human epithelial cells. Cell. Microbiol. 1:237-247. - PubMed
-
- Al-Younes, H. M., T. Rudel, V. Brinkmann, A. J. Szczepek, and T. F. Meyer. 2001. Low iron availability modulates the course of Chlamydia pneumoniae infection. Cell. Microbiol. 3:427-437. - PubMed
-
- Avruch, J., C. Belham, Q. Wenig, K. Hara, and K. Yonezawa. 2001. The p70 S6 kinase integrates nutrient and growth signals to control translational capacity. Prog. Mol. Subcell. Biol. 26:115-154. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

