Chromatin landscape dynamics of the Il4-Il13 locus during T helper 1 and 2 development
- PMID: 15272080
- PMCID: PMC509214
- DOI: 10.1073/pnas.0403334101
Chromatin landscape dynamics of the Il4-Il13 locus during T helper 1 and 2 development
Abstract
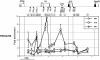
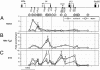
Il4 and Il13 encode the canonical T helper 2 (TH2) cytokines responsible both for promoting immune responses against extracellular pathogens and, when misregulated, causing allergic and autoimmune disease. The expression potential of these genes undergoes developmentally programmed repression and enhancement during commitment of naïve CD4+ T cells to the mature T helper 1 (TH1) and TH2 fates, respectively. Thus, like the globin locus, the TH2 cytokine locus provides a highly tractable system to study a developmental fate choice leading to alternative transcriptional states of either silence or permissivity. We used quantitative chromatin immunoprecipitation and RT-PCR to correlate changes in the transcriptional states of Il4 and Il13 with markers of permissive chromatin across the Il4-Il13 locus in naïve CD4+ T cells undergoing TH1 and TH2 differentiation. We provide evidence that DNaseI hypersensitive site V in the Il4 3' enhancer is the likely target for signals maintaining Il4 and Il13 transcriptional permissivity in naïve cells. We also demonstrate rapid acquisition of differences in H3 acetylation between TH1- and TH2-primed cells, indicating a developmentally early role for cytokine signaling in the process of TH cell fate determination. Finally, we show that transcriptional repression correlates with the disappearance of permissive H3 modifications from everywhere in the Il4-Il13 locus except hypersensitive site IV, suggesting a critical role for this element in the maintenance of transcriptional repression. Our findings are consistent with a progressive regulatory element activation/deactivation model of TH1/TH2 development.
Figures



Similar articles
-
Intergenic transcription is not required in Th2 cells to maintain histone acetylation and transcriptional permissiveness at the Il4-Il13 locus.J Immunol. 2005 Dec 15;175(12):8146-53. doi: 10.4049/jimmunol.175.12.8146. J Immunol. 2005. PMID: 16339553
-
EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells.J Biol Chem. 2005 Sep 9;280(36):31470-7. doi: 10.1074/jbc.M504766200. Epub 2005 Jul 11. J Biol Chem. 2005. PMID: 16009709
-
Regulation of Th2 differentiation and Il4 locus accessibility.Annu Rev Immunol. 2006;24:607-56. doi: 10.1146/annurev.immunol.23.021704.115821. Annu Rev Immunol. 2006. PMID: 16551261 Review.
-
Probabilistic regulation in TH2 cells accounts for monoallelic expression of IL-4 and IL-13.Immunity. 2005 Jul;23(1):89-99. doi: 10.1016/j.immuni.2005.05.008. Immunity. 2005. PMID: 16039582
-
Chromatin remodeling and T helper subset differentiation.IUBMB Life. 2000 Jun;49(6):473-8. doi: 10.1080/15216540050166990. IUBMB Life. 2000. PMID: 11032239 Review.
Cited by
-
DNA methylation-histone modification relationships across the desmin locus in human primary cells.BMC Mol Biol. 2009 May 27;10:51. doi: 10.1186/1471-2199-10-51. BMC Mol Biol. 2009. PMID: 19473514 Free PMC article.
-
Regulation of the Il4 gene is independently controlled by proximal and distal 3' enhancers in mast cells and basophils.Mol Cell Biol. 2007 Dec;27(23):8087-97. doi: 10.1128/MCB.00631-07. Epub 2007 Oct 1. Mol Cell Biol. 2007. PMID: 17908791 Free PMC article.
-
Epigenetic Reprogramming of CD4+ Helper T Cells as a Strategy to Improve Anticancer Immunotherapy.Front Immunol. 2021 Jun 28;12:669992. doi: 10.3389/fimmu.2021.669992. eCollection 2021. Front Immunol. 2021. PMID: 34262562 Free PMC article. Review.
-
Control of IL-4 expression in T helper 1 and 2 cells.Immunology. 2008 Aug;124(4):437-44. doi: 10.1111/j.1365-2567.2008.02845.x. Epub 2008 May 9. Immunology. 2008. PMID: 18479352 Free PMC article. Review.
-
In vitro and in vivo differentiated effector CD8 T cells display divergent histone acetylation patterns within the Ifng locus.Immunol Lett. 2009 Feb 21;122(2):214-8. doi: 10.1016/j.imlet.2009.01.001. Epub 2009 Feb 3. Immunol Lett. 2009. PMID: 19195486 Free PMC article.
References
-
- Reiner, S. & Locksley, R. (1995) The Regulation of Immunity to Leishmania Major (Annual Reviews, Palo Alto). - PubMed
-
- Urban, J. F., Jr., Madden, K. B., Svetic, A., Cheever, A., Trotta, P. P., Gause, W. C., Katona, I. M. & Finkelman, F. D. (1992) Immunol. Rev. 127, 205–220. - PubMed
-
- Takemoto, N., Koyano-Nakagawa, N., Yokota, T., Arai, N., Miyatake, S. & Arai, K. (1998) Int. Immunol. 10, 1981–1985. - PubMed
-
- Agarwal, S. & Rao, A. (1998) Immunity 9, 765–775. - PubMed
-
- Lee, G. R., Fields, P. E., Griffin, T. J. & Flavell, R. A. (2003) Immunity 19, 145–153. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

