Novel polymorphisms in mec genes and a new mec complex type in methicillin-resistant Staphylococcus aureus isolates obtained in rural Wisconsin
- PMID: 15273123
- PMCID: PMC478485
- DOI: 10.1128/AAC.48.8.3080-3085.2004
Novel polymorphisms in mec genes and a new mec complex type in methicillin-resistant Staphylococcus aureus isolates obtained in rural Wisconsin
Abstract
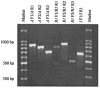
We determined allelic polymorphisms in the mec complexes of 524 methicillin-resistant Staphylococcus aureus isolates by partial or complete sequencing of three mec genes, mecA, mecI, and mecR1. The isolates had been collected over a 10-year period from patients living in rural Wisconsin, where the use of antibiotics was expected to be lower than in the bigger cities. Of the 18 mutation types identified, 16 had not been described previously. The five most common mutations were a mutation 7 nucleotides (nt) upstream from the start site (G-->T) in the mecA promoter (34.7%), an E246G change encoded by mecA (2.2%), a cytosine insertion at codon 257 in mecA (13.2%), an N121K change encoded by mecI (7.8%), and a major mecI-mecR1 deletion designated as a class B1 mec complex deletion type (25.4%). There was a high degree of conservation of the amino acid sequence of MecR1. Strains with the same mutations had variable resistance to oxacillin, and the median MIC for isolates that harbored the 7-nt-upstream mutation was lower than that for strains harboring other mutations. Our data suggest that the mecA promoter mutation plays a more important role in determining methicillin resistance than mutations in mecI and mecA do. Eighty-five percent of the tested isolates (n = 148) with the mecA promoter mutation and the class B1 mec complex deletion belonged to the same major clonal group, identified as MCG-2, and harbored the type IV staphylococcal cassette chromosome mec DNA. There was also a wide range of oxacillin MICs for strains with wild-type mecA, mecI, and mecR1 sequences, suggesting that the genetic backgrounds of clinical strains are significant in determining susceptibility to methicillin.
Figures


References
-
- Archer, G. L., and J. M. Bosilevac. 2001. Signaling antibiotic resistance in staphylococci. Science 291:1915-1916. - PubMed
-
- Berger-Bachi, B., and S. Rohrer. 2002. Factors influencing methicillin resistance in staphylococci. Arch. Microbiol. 178:165-171. - PubMed
-
- Chambers, H. F., and M. Sachdeva. 1990. Binding of beta-lactam antibiotics to penicillin-binding proteins in methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 161:1170-1176. - PubMed
-
- Hiramatsu, K. 1995. Molecular evolution of MRSA. Microbiol. Immunol. 39:531-543. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

