Sequence-specific artificial ribonucleases. I. Bis-imidazole-containing oligonucleotide conjugates prepared using precursor-based strategy
- PMID: 15273275
- PMCID: PMC506794
- DOI: 10.1093/nar/gkh702
Sequence-specific artificial ribonucleases. I. Bis-imidazole-containing oligonucleotide conjugates prepared using precursor-based strategy
Abstract
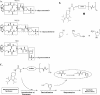
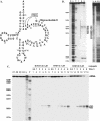
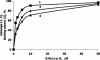
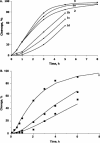
Antisense oligonucleotide conjugates, bearing constructs with two imidazole residues, were synthesized using a precursor-based technique employing post-synthetic histamine functionalization of oligonucleotides bearing methoxyoxalamido precursors at the 5'-termini. The conjugates were assessed in terms of their cleavage activities using both biochemical assays and conformational analysis by molecular modelling. The oligonucleotide part of the conjugates was complementary to the T-arm of yeast tRNA(Phe) (44-60 nt) and was expected to deliver imidazole groups near the fragile sequence C61-ACA-G65 of the tRNA. The conjugates showed ribonuclease activity at neutral pH and physiological temperature resulting in complete cleavage of the target RNA, mainly at the C63-A64 phosphodiester bond. For some constructs, cleavage was completed within 1-2 h under optimal conditions. Molecular modelling was used to determine the preferred orientation(s) of the cleaving group(s) in the complexes of the conjugates with RNA target. Cleaving constructs bearing two imidazole residues were found to be conformationally highly flexible, adopting no preferred specific conformation. No interactions other than complementary base pairing between the conjugates and the target were found to be the factors stabilizing the 'active' cleaving conformation(s).
Figures
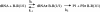
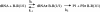

Similar articles
-
[Highly efficient site-directed RNA cleavage by imidazole-containing conjugates of antisense oligonucleotides].Mol Biol (Mosk). 2002 Jul-Aug;36(4):731-9. Mol Biol (Mosk). 2002. PMID: 12173480 Russian.
-
Sequence-specific cleavage of yeast tRNA(Phe) with oligonucleotides conjugated to a diimidazole construct.Antisense Nucleic Acid Drug Dev. 1997 Feb;7(1):39-42. doi: 10.1089/oli.1.1997.7.39. Antisense Nucleic Acid Drug Dev. 1997. PMID: 9055037
-
Cleavage of yeast tRNAPhe with complementary oligonucleotide conjugated to a small ribonuclease mimic.FEBS Lett. 2000 Sep 22;481(3):277-80. doi: 10.1016/s0014-5793(00)02029-9. FEBS Lett. 2000. PMID: 11007978
-
The sequence-specific cleavage of RNA by artificial chemical ribonucleases.Antisense Nucleic Acid Drug Dev. 1997 Aug;7(4):423-30. doi: 10.1089/oli.1.1997.7.423. Antisense Nucleic Acid Drug Dev. 1997. PMID: 9303194 Review.
-
Discovering antisense reagents by hybridization of RNA to oligonucleotide arrays.Ciba Found Symp. 1997;209:38-44; discussion 44-6. doi: 10.1002/9780470515396.ch4. Ciba Found Symp. 1997. PMID: 9383567 Review.
Cited by
-
Site-Selective Artificial Ribonucleases: Renaissance of Oligonucleotide Conjugates for Irreversible Cleavage of RNA Sequences.Molecules. 2021 Mar 19;26(6):1732. doi: 10.3390/molecules26061732. Molecules. 2021. PMID: 33808835 Free PMC article. Review.
-
Targeted inhibition of the hepatitis C internal ribosomal entry site genomic RNA with oligonucleotide conjugates.Nucleic Acids Res. 2007;35(20):6778-87. doi: 10.1093/nar/gkm770. Epub 2007 Oct 5. Nucleic Acids Res. 2007. PMID: 17921501 Free PMC article.
-
Synthesis of functionalised nucleosides for incorporation into nucleic acid-based serine protease mimics.Molecules. 2007 Jan 31;12(1):114-29. doi: 10.3390/12010114. Molecules. 2007. PMID: 17693958 Free PMC article.
-
Cleavage of pyrene-stabilized RNA bulge loops by trans-(±)-cyclohexane-1,2-diamine.Chem Cent J. 2012 Jan 13;6:3. doi: 10.1186/1752-153X-6-3. Chem Cent J. 2012. PMID: 22244351 Free PMC article.
-
Substrate specificity and kinetic framework of a DNAzyme with an expanded chemical repertoire: a putative RNaseA mimic that catalyzes RNA hydrolysis independent of a divalent metal cation.Nucleic Acids Res. 2004 Dec 29;32(22):6660-72. doi: 10.1093/nar/gkh1007. Print 2004. Nucleic Acids Res. 2004. PMID: 15625232 Free PMC article.
References
-
- Stern M.K., Bashkin,J.K. and Sall,E.D. (1990) Hydrolysis of RNA by transition metal complexes. J. Am. Chem. Soc., 112, 5357–5359.
-
- Barbier B. and Brack,A. (1987) Search for catalytic properties of simple polypeptides. Orig. Life Evol. Biosph., 17, 381–390. - PubMed
-
- Li Y., Zhao,Y., Hatfield,S., Wan,R., Zhu,Q., Li,X., McMills,M., Ma,Y., Li,J., Brown,K.L., He,C., Liu,F. and Chen,X. (2000) Dipeptide seryl-histidine and related oligopeptides cleave DNA, protein and carboxyl ester. Bioorg. Med. Chem., 8, 2675–2680. - PubMed
-
- Michaelis K. and Kalesse,M. (1999) Selective cleavage of the HIV-1 TAR–RNA with a peptide-cyclen conjugate. Angew. Chem. Int. Ed. Engl., 38, 2243–2245. - PubMed
-
- Yoschinari K., Yamazaki,K. and Komiyama,M. (1991) Oligoamines as a simple and efficient catalysts. J. Am. Chem. Soc., 113, 5899–5901.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

