Molecular-timetable methods for detection of body time and rhythm disorders from single-time-point genome-wide expression profiles
- PMID: 15273285
- PMCID: PMC509173
- DOI: 10.1073/pnas.0401882101
Molecular-timetable methods for detection of body time and rhythm disorders from single-time-point genome-wide expression profiles
Abstract
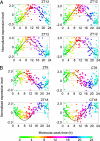
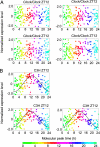
Detection of individual body time (BT) via a single-time-point assay has been a longstanding unfulfilled dream in medicine, because BT information can be exploited to maximize potency and minimize toxicity during drug administration and thus will enable highly optimized medication. To achieve this dream, we created a "molecular timetable" composed of >100 "time-indicating genes," whose gene expression levels can represent internal BT. Here we describe a robust method called the "molecular-timetable method" for BT detection from a single-time-point expression profile. The power of this method is demonstrated by the sensitive and accurate detection of BT and the sensitive diagnosis of rhythm disorders. These results demonstrate the feasibility of BT detection based on single-time-point sampling, suggest the potential for expression-based diagnosis of rhythm disorders, and may translate functional genomics into chronotherapy and personalized medicine.
Figures

References
-
- Reppert, S. M. & Weaver, D. R. (2002) Nature 418, 935–941. - PubMed
-
- Akhtar, R. A., Reddy, A. B., Maywood, E. S., Clayton, J. D., King, V. M., Smith, A. G., Gant, T. W., Hastings, M. H. & Kyriacou, C. P. (2002) Curr. Biol. 12, 540–550. - PubMed
-
- Panda, S., Antoch, M. P., Miller, B. H., Su, A. I., Schook, A. B., Straume, M., Schultz, P. G., Kay, S. A., Takahashi, J. S. & Hogenesch, J. B. (2002) Cell 109, 307–320. - PubMed
-
- Storch, K. F., Lipan, O., Leykin, I., Viswanathan, N., Davis, F. C., Wong, W. H. & Weitz, C. J. (2002) Nature 417, 78–83. - PubMed
-
- Ueda, H. R., Chen, W., Adachi, A., Wakamatsu, H., Hayashi, S., Takasugi, T., Nagano, M., Nakahama, K., Suzuki, Y., Sugano, S., et al. (2002) Nature 418, 534–539. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

