Watching the photosynthetic apparatus in native membranes
- PMID: 15273291
- PMCID: PMC509197
- DOI: 10.1073/pnas.0404350101
Watching the photosynthetic apparatus in native membranes
Abstract
Over the last 9 years, the structures of the various components of the bacterial photosynthetic apparatus or their homologues have been determined by x-ray crystallography to at least 4.8-A resolution. Despite this wealth of structural information on the individual proteins, there remains an urgent need to examine the architecture of the photosynthetic apparatus in intact photosynthetic membranes. Information on the arrangement of the different complexes in a native system will help us to understand the processes that ensure the remarkably high quantum efficiency of the system. In this work we report images obtained with an atomic force microscope of native photosynthetic membranes from the bacterium Rhodospirillum photometricum. Several proteins can be seen and identified at molecular resolution, allowing the analysis and modeling of the lateral organization of multiple components of the photosynthetic apparatus within a native membrane. Analysis of the distribution of the complexes shows that their arrangement is far from random, with significant clustering both of antenna complexes and core complexes. The functional significance of the observed distribution is discussed.
Figures
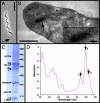
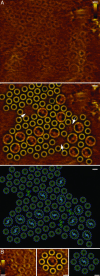

References
-
- Bibby, T., Nield, J., Partensky, F. & Barber, J. (2001) Nature 413, 590. - PubMed
-
- Abrahams, J. P., Leslie, A. G. W., Lutter, R. & Walker, J. E. (1994) Nature 370, 621–628. - PubMed
-
- Kühlbrandt, W. (1995) Nature 374, 497.
-
- Hu, X., Ritz, T., Damjanovic, A., Autenrieth, F. & Schulten, K. (2002) Q. Rev. Biophys. 35, 1–62. - PubMed
-
- Deisenhofer, J., Epp, O., Miki, K., Huber, R. & Michel, H. (1984) J. Mol. Biol. 180, 385–398. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

