The Ustilago maydis a2 mating-type locus genes lga2 and rga2 compromise pathogenicity in the absence of the mitochondrial p32 family protein Mrb1
- PMID: 15273296
- PMCID: PMC519210
- DOI: 10.1105/tpc.104.022657
The Ustilago maydis a2 mating-type locus genes lga2 and rga2 compromise pathogenicity in the absence of the mitochondrial p32 family protein Mrb1
Abstract
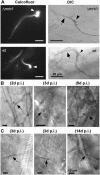
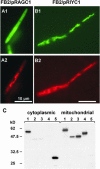
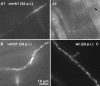
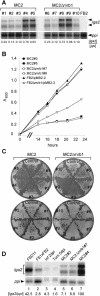
The Ustilago maydis mrb1 gene specifies a mitochondrial matrix protein with significant similarity to mitochondrial p32 family proteins known from human and many other eukaryotic species. Compatible mrb1 mutant strains were able to mate and form dikaryotic hyphae; however, proliferation within infected tissue and the ability to induce tumor development of infected maize (Zea mays) plants were drastically impaired. Surprisingly, manifestation of the mrb1 mutant phenotype selectively depended on the a2 mating type locus. The a2 locus contains, in addition to pheromone signaling components, the genes lga2 and rga2 of unknown function. Deletion of lga2 in an a2Deltamrb1 strain fully restored pathogenicity, whereas pathogenicity was partially regained in an a2Deltamrb1Deltarga2 strain, implicating a concerted action between Lga2 and Rga2 in compromising pathogenicity in Deltamrb1 strains. Lga2 and Rga2 localized to mitochondria and Mrb1 interacted with Rga2 in the yeast two-hybrid system. Conditional expression of lga2 in haploid cells reduced vegetative growth, conferred mitochondrial fragmentation and mitochondrial DNA degradation, and interfered with respiratory activity. The consequences of lga2 overexpression depended on the expression strength and were greatly exacerbated in Deltamrb1 mutants. We propose that Lga2 interferes with mitochondrial fusion and that Mrb1 controls this activity, emphasizing a critical link between mitochondrial morphology and pathogenicity.
Figures




References
-
- Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J.W., and Elledge, S.J. (1996). SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86, 263–274. - PubMed
-
- Banuett, F., and Herskowitz, I. (1996). Discrete developmental stages during teliospore formation in the corn smut fungus, Ustlago maydis. Development 122, 2965–2976. - PubMed
-
- Basse, C.W., Kolb, S., and Kahmann, R. (2002). A maize-specifically expressed gene cluster in Ustilago maydis. Mol. Microbiol. 43, 75–93. - PubMed
-
- Basse, C.W., and Steinberg, G. (2004). Ustilago maydis, model system for analysis of the molecular basis of fungal pathogenicity. Mol. Plant. Pathol. 5, 83–92. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

