Evolutionary constraints associated with functional specificity of the CMGC protein kinases MAPK, CDK, GSK, SRPK, DYRK, and CK2alpha
- PMID: 15273306
- PMCID: PMC2279817
- DOI: 10.1110/ps.04637904
Evolutionary constraints associated with functional specificity of the CMGC protein kinases MAPK, CDK, GSK, SRPK, DYRK, and CK2alpha
Abstract
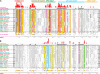
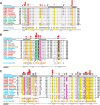
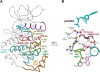
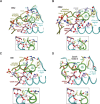
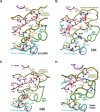
Amino acid residues associated with functional specificity of cyclin-dependent kinases (CDKs), mitogen-activated protein kinases (MAPKs), glycogen synthase kinases (GSKs), and CDK-like kinases (CLKs), which are collectively termed the CMGC group, were identified by categorizing and quantifying the selective constraints acting upon these proteins during evolution. Many constraints specific to CMGC kinases correspond to residues between the N-terminal end of the activation segment and a CMGC-conserved insert segment associated with coprotein binding. The strongest such constraint is imposed on a "CMGC-arginine" near the substrate phosphorylation site with a side chain that plays a role both in substrate recognition and in kinase activation. Two nearby buried waters, which are also present in non-CMGC kinases, typically position the main chain of this arginine relative to the catalytic loop. These and other CMGC-specific features suggest a structural linkage between coprotein binding, substrate recognition, and kinase activation. Constraints specific to individual subfamilies point to mechanisms for CMGC kinase specialization. Within casein kinase 2alpha (CK2alpha), for example, the binding of one of the buried waters appears prohibited by the side chain of a leucine that is highly conserved within CK2alpha and that, along with substitution of lysine for the CMGC-arginine, may contribute to the broad substrate specificity of CK2alpha by relaxing characteristically conserved, precise interactions near the active site. This leucine is replaced by a conserved isoleucine or valine in other CMGC kinases, thereby illustrating the potential functional significance of subtle amino acid substitutions. Analysis of other CMGC kinases similarly suggests candidate family-specific residues for experimental follow-up.
Figures



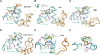
Similar articles
-
Evolved to be active: sulfate ions define substrate recognition sites of CK2alpha and emphasise its exceptional role within the CMGC family of eukaryotic protein kinases.J Mol Biol. 2007 Jul 13;370(3):427-38. doi: 10.1016/j.jmb.2007.04.068. Epub 2007 May 5. J Mol Biol. 2007. PMID: 17524418
-
Structural and evolutionary divergence of eukaryotic protein kinases in Apicomplexa.BMC Evol Biol. 2011 Nov 2;11:321. doi: 10.1186/1471-2148-11-321. BMC Evol Biol. 2011. PMID: 22047078 Free PMC article.
-
The crystal structure of the protein kinase HIPK2 reveals a unique architecture of its CMGC-insert region.J Biol Chem. 2019 Sep 13;294(37):13545-13559. doi: 10.1074/jbc.RA119.009725. Epub 2019 Jul 24. J Biol Chem. 2019. PMID: 31341017 Free PMC article.
-
CMGC Kinases in Health and Cancer.Cancers (Basel). 2023 Jul 28;15(15):3838. doi: 10.3390/cancers15153838. Cancers (Basel). 2023. PMID: 37568654 Free PMC article. Review.
-
Interplay Between CMGC Kinases Targeting SR Proteins and Viral Replication: Splicing and Beyond.Front Microbiol. 2021 Mar 29;12:658721. doi: 10.3389/fmicb.2021.658721. eCollection 2021. Front Microbiol. 2021. PMID: 33854493 Free PMC article. Review.
Cited by
-
Co-conserved MAPK features couple D-domain docking groove to distal allosteric sites via the C-terminal flanking tail.PLoS One. 2015 Mar 23;10(3):e0119636. doi: 10.1371/journal.pone.0119636. eCollection 2015. PLoS One. 2015. PMID: 25799139 Free PMC article.
-
Structural and Enzymological Evidence for an Altered Substrate Specificity in Okur-Chung Neurodevelopmental Syndrome Mutant CK2αLys198Arg.Front Mol Biosci. 2022 Apr 4;9:831693. doi: 10.3389/fmolb.2022.831693. eCollection 2022. Front Mol Biosci. 2022. PMID: 35445078 Free PMC article.
-
Functional flexibility of human cyclin-dependent kinase-2 and its evolutionary conservation.Protein Sci. 2008 Jan;17(1):22-33. doi: 10.1110/ps.072951208. Epub 2007 Nov 27. Protein Sci. 2008. PMID: 18042686 Free PMC article.
-
SUMOylation of the Forkhead transcription factor FOXL2 promotes its stabilization/activation through transient recruitment to PML bodies.PLoS One. 2011;6(10):e25463. doi: 10.1371/journal.pone.0025463. Epub 2011 Oct 12. PLoS One. 2011. PMID: 22022399 Free PMC article.
-
Phosphorylation of the transcription activator CLOCK regulates progression through a ∼ 24-h feedback loop to influence the circadian period in Drosophila.J Biol Chem. 2014 Jul 11;289(28):19681-93. doi: 10.1074/jbc.M114.568493. Epub 2014 May 28. J Biol Chem. 2014. PMID: 24872414 Free PMC article.
References
-
- Adams, J.A. 2003. Activation loop phosphorylation and catalysis in protein kinases: Is there functional evidence for the autoinhibitor model? Biochemistry 42 601–607. - PubMed
-
- Battistutta, R., Sarno, S., De Moliner, E., Marin, O., Issinger, O.G., Zanotti, G., and Pinna, L.A. 2000. The crystal structure of the complex of Zea mays α subunit with a fragment of human β subunit provides the clue to the architecture of protein kinase CK2 holoenzyme. Eur. J. Biochem. 267 5184–5190. - PubMed
-
- Bax, B., Carter, P.S., Lewis, C., Guy, A.R., Bridges, A., Tanner, R., Pettman, G., Mannix, C., Culbert, A.A., Brown, M.J., et al. 2001. The structure of phosphorylated GSK-3β complexed with a peptide, FRATtide, that inhibits β-catenin phosphorylation. Structure 9 1143–1152. - PubMed
-
- Becker, W. and Joost, H.G. 1999. Structural and functional characteristics of DYRK, a novel subfamily of protein kinases with dual specificity. Prog. Nucleic Acid Res. Mol. Biol. 62 1–17. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

