Solution structure of an RNA stem-loop derived from the 3' conserved region of eel LINE UnaL2
- PMID: 15273327
- PMCID: PMC1370625
- DOI: 10.1261/rna.7460104
Solution structure of an RNA stem-loop derived from the 3' conserved region of eel LINE UnaL2
Abstract
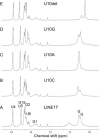
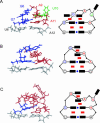
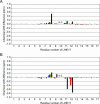
The eel long interspersed element (LINE) UnaL2 and its partner short interspersed element (SINE) share a conserved 3' tail containing a stem-loop that is critical for their retrotransposition. Presumably, the first step of retrotransposition is the recognition of their 3' tails by UnaL2-encoded reverse transcriptase. The solution structure of a 17-nucleotide RNA derived from the 3' tail of UnaL2 was determined by NMR. The GGAUA loop forms a specific structure in which the uridine is exposed to solvent with the third and fifth adenosines stacked. A sharp turn in the phosphodiester backbone occurs between the second guanosine and third adenosine. When the uridine is mutated (but not deleted), all mutants form the loop structure, indicating that the loop structure requires an exposed fourth residue. The retrotransposition assay in HeLa cells revealed that retrotransposition requires the second guanosine, although any nucleoside functions at the fourth position, suggesting that UnaL2 reverse transcriptase specifically recognizes the 5' side of the GGANA loop.
Figures

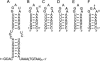



References
-
- Allain, F.H. and Varani, G. 1995. Structure of the P1 helix from group I self-splicing introns. J. Mol. Biol. 250: 333–353. - PubMed
-
- Altona, C. and Sundaralingam, M. 1973. Conformational analysis of the sugar ring in nucleosides and nucleotides. Improved method for the interpretation of proton magnetic resonance coupling constants. J. Am. Chem. Soc. 95: 2333–2344. - PubMed
-
- Boeke, J.D. and Devine, S.E. 1998. Yeast retrotransposons: Finding a nice quiet neighborhood. Cell 93: 1087–1089. - PubMed
-
- Deininger, P.L. and Batzer, M.A. 1993. Evolution of retroposons. Evolutionary Biol. 27: 157–196.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
