E. coli trp repressor forms a domain-swapped array in aqueous alcohol
- PMID: 15274929
- PMCID: PMC3228604
- DOI: 10.1016/j.str.2004.03.019
E. coli trp repressor forms a domain-swapped array in aqueous alcohol
Abstract
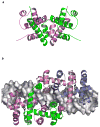
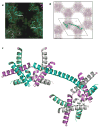
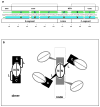
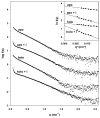
The E. coli trp repressor (trpR) homodimer recognizes its palindromic DNA binding site through a pair of flexible helix-turn-helix (HTH) motifs displayed on an intertwined helical core. Flexible N-terminal arms mediate association between dimers bound to tandem DNA sites. The 2.5 A X-ray structure of trpR crystallized in 30% (v/v) isopropanol reveals a substantial conformational rearrangement of HTH motifs and N-terminal arms, with the protein appearing in the unusual form of an ordered 3D domain-swapped supramolecular array. Small angle X-ray scattering measurements show that the self-association properties of trpR in solution are fundamentally altered by isopropanol.
Figures



References
-
- Abrahams JP, Leslie AGW. Methods used in the structure determination of bovine mitochondrial F-1 ATPase. 5. 1996;2:30–42. - PubMed
-
- Babu KR, Moradian A, Douglas DJ. The methanol-induced conformational transitions of beta-lactoglobulin, cytochrome c, and ubiquitin at low pH: a study by electrospray ionization mass spectrometry. J Am Soc Mass Spectrom. 2001;12:317–328. - PubMed
-
- Buck M. Trifluoroethanol and colleagues: cosolvents come of age. Recent studies with peptides and proteins. Q Rev Biophys. 1998;31:297–355. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

