Differential gene expression in ovarian carcinoma: identification of potential biomarkers
- PMID: 15277215
- PMCID: PMC1618570
- DOI: 10.1016/S0002-9440(10)63306-8
Differential gene expression in ovarian carcinoma: identification of potential biomarkers
Abstract
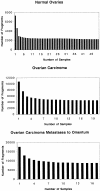
Ovarian cancer remains the fifth leading cause of cancer death for women in the United States. In this study, the gene expression of 20 ovarian carcinomas, 17 ovarian carcinomas metastatic to the omentum, and 50 normal ovaries was determined by Gene Logic Inc. using Affymetrix GeneChip HU_95 arrays containing approximately 12,000 known genes. Differences in gene expression were quantified as fold changes in gene expression in ovarian carcinomas compared to normal ovaries and ovarian carcinoma metastases. Genes up-regulated in ovarian carcinoma tissue samples compared to more than 300 other normal and diseased tissue samples were identified. Seven genes were selected for further screening by immunohistochemistry to determine the presence and localization of the proteins. These seven genes were: the beta8 integrin subunit, bone morphogenetic protein-7, claudin-4, collagen type IX alpha2, cellular retinoic acid binding protein-1, forkhead box J1, and S100 calcium-binding protein A1. Statistical analyses showed that the beta8 integrin subunit, claudin-4, and S100A1 provided the best distinction between ovarian carcinoma and normal ovary tissues, and may serve as the best candidate tumor markers among the seven genes studied. These results suggest that further exploration into other up-regulated genes may identify novel diagnostic, therapeutic, and/or prognostic biomarkers in ovarian carcinoma.
Figures



References
-
- Ahmedin J, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ. Cancer statistics. CA Cancer J Clin. 2003;53:5–26. - PubMed
-
- Verheijen RHM, von Mensdorff-Pouilly S, van Kamp GJ, Kenemans P. CA 125: fundamental and clinical aspects. Cancer Biol. 1999;9:117–124. - PubMed
-
- Bast Jr RC, Urban N, Shridhar V, Smith D, Zhang Z, Skates S, Lu K, Liu J, Fishman D, Mills G: Early detection of ovarian cancer: promise and reality. Ovarian Cancer. Edited by Stack MS, Fishman DA. Cancer Treatment and Research, vol 107 Edited by Rosen ST (Series Editor). Boston, Kluwer Academic Publishers, 2002, pp 61–97 - PubMed
-
- Welsh JB, Zarrinkar PP, Sapinoso LM, Kern SG, Behling CA, Monk BJ, Lockhart DJ, Burger RA, Hampton GM. Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc Natl Acad Sci USA. 2001;98:1176–1181. - PMC - PubMed
-
- Hough CD, Cho KR, Zonderman AB, Schwartz DR, Morin PJ. Coordinately up-regulated genes in ovarian cancer. Cancer Res. 2001;61:3869–3876. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

