Integrin beta3 regions controlling binding of murine mAb 7E3: implications for the mechanism of integrin alphaIIbbeta3 activation
- PMID: 15277669
- PMCID: PMC516524
- DOI: 10.1073/pnas.0404201101
Integrin beta3 regions controlling binding of murine mAb 7E3: implications for the mechanism of integrin alphaIIbbeta3 activation
Abstract
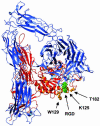
Abciximab, a derivative of the murine mAb 7E3, protects against ischemic complications of percutaneous coronary interventions by inhibiting ligand binding to the alphaIIbbeta3 receptor. In this study we identified regions on integrin beta3 that control 7E3 binding. Murine/human amino acid substitutions were created in two regions of the betaA domain that previous studies found to influence 7E3 binding: the C177-C184 loop and K125-N133. The T182N substitution and a K125Q mutation reduced 7E3 binding to human beta3 in complex with alphaIIb. The introduction of both the human C177-C184 region and human W129 into murine beta3 was necessary and sufficient to permit 7E3 binding to the human alphaIIb/murine beta3 complex. Although we cannot exclude allosteric effects, we propose that 7E3 binds between C177-C184 and W129, which are within 15 A of each other in the crystal structure and close to the beta3 metal ion-dependent adhesion site. We previously demonstrated that 7E3 binds more rapidly to activated than unactivated platelets. Because it has been proposed that alphaIIbbeta3 changes from a bent to an extended conformation upon activation, we hypothesized that 7E3 binds less well to the bent than the extended conformation. In support of this hypothesis we found that 7E3 bound less well to an alphaIIbbeta3 construct locked in a bent conformation, and unlocking the conformation restored 7E3 binding. Thus, our data are consistent with alphaIIbbeta3 existing in variably bent conformations in equilibrium with each other on unactivated platelets, and activation resulting in alphaIIbbeta3 adopting a more extended conformation.
Figures
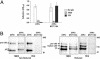

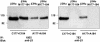
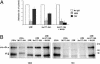


Similar articles
-
Electron microscopy shows that binding of monoclonal antibody PT25-2 primes integrin αIIbβ3 for ligand binding.Blood Adv. 2021 Apr 13;5(7):1781-1790. doi: 10.1182/bloodadvances.2020004166. Blood Adv. 2021. PMID: 33760023 Free PMC article.
-
Structure and function of murine alphaIIbbeta3 (GPIIb/IIIa): studies using monoclonal antibodies and beta3-null mice,Thromb Haemost. 2000 Dec;84(6):1103-8. Thromb Haemost. 2000. PMID: 11154120
-
An αIIbβ3 monoclonal antibody traps a semiextended conformation and allosterically inhibits large ligand binding.Blood Adv. 2024 Aug 27;8(16):4398-4409. doi: 10.1182/bloodadvances.2024013177. Blood Adv. 2024. PMID: 38968144 Free PMC article.
-
Beta3 tyrosine phosphorylation in alphaIIbbeta3 (platelet membrane GP IIb-IIIa) outside-in integrin signaling.Thromb Haemost. 2001 Jul;86(1):246-58. Thromb Haemost. 2001. PMID: 11487013 Review.
-
Cytoplasmic binding partners of the platelet integrin alphaIIb beta3.Front Biosci. 2007 Jan 1;12:2038-49. doi: 10.2741/2209. Front Biosci. 2007. PMID: 17127442 Review.
Cited by
-
Volatile anesthetics, not intravenous anesthetic propofol bind to and attenuate the activation of platelet receptor integrin αIIbβ3.PLoS One. 2013;8(4):e60415. doi: 10.1371/journal.pone.0060415. Epub 2013 Apr 3. PLoS One. 2013. PMID: 23573252 Free PMC article.
-
Elevating local concentrations of GPIIb-IIIa antagonists counteracts platelet thrombus stability.J Thromb Thrombolysis. 2013 Jul;36(1):31-41. doi: 10.1007/s11239-012-0814-7. J Thromb Thrombolysis. 2013. PMID: 23073747 Free PMC article.
-
Integrins αvβ3 and α4β1 act as coreceptors for fractalkine, and the integrin-binding defective mutant of fractalkine is an antagonist of CX3CR1.J Immunol. 2012 Dec 15;189(12):5809-19. doi: 10.4049/jimmunol.1200889. Epub 2012 Nov 2. J Immunol. 2012. PMID: 23125415 Free PMC article.
-
Platelet binding to polymerizing fibrin is avidity driven and requires activated αIIbβ3 but not fibrin cross-linking.Blood Adv. 2021 Oct 26;5(20):3986-4002. doi: 10.1182/bloodadvances.2021005142. Blood Adv. 2021. PMID: 34647980 Free PMC article.
-
The heparin-binding domain of VEGF165 directly binds to integrin αvβ3 and VEGFR2/KDR D1: a potential mechanism of negative regulation of VEGF165 signaling by αvβ3.Front Cell Dev Biol. 2024 May 9;12:1347616. doi: 10.3389/fcell.2024.1347616. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38803393 Free PMC article.
References
-
- Humphries, M. J. (2000) Biochem. Soc. Trans. 28, 311–339. - PubMed
-
- Hynes, R. (2002) Cell 110, 673–687. - PubMed
-
- Charo, I. F., Bekeart, L. S. & Phillips, D. R. (1987) J. Biol. Chem. 262, 9935–9938. - PubMed
-
- Coller, B. S., Peerschke, E. I., Seligsohn, U., Scudder, L. E., Nurden, A. T. & Rosa, J. P. (1986) J. Lab. Clin. Med. 107, 384–392. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

