The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor
- PMID: 15277670
- PMCID: PMC509221
- DOI: 10.1073/pnas.0404220101
The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor
Abstract
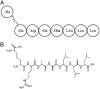
SapB is a morphogenetic peptide that is important for aerial mycelium formation by the filamentous bacterium Streptomyces coelicolor. Production of SapB commences during aerial mycelium formation and depends on most of the genes known to be required for the morphogenesis of aerial hyphae. Furthermore, the application of purified SapB to mutants blocked in morphogenesis restores their capacity to form aerial hyphae. Here, we present evidence that SapB is a lantibiotic-like peptide that is derived by posttranslational modification from the product of a gene (ramS) in the four-gene ram operon, which is under the control of the regulatory gene ramR. We show that the product of another gene in the operon (ramC) contains a region that is similar to enzymes involved in the biosynthesis of lantibiotics, suggesting that it might be involved in the posttranslational processing of RamS. We conclude that SapB is derived from RamS through proteolytic cleavage and the introduction of four dehydroalanine residues and two lanthionine bridges. We provide an example of a morphogenetic role for an antibiotic-like molecule.
Figures




References
-
- Kelemen, G. H. & Buttner, M. J. (1998) Curr. Opin. Microbiol. 1, 656-662. - PubMed
-
- Chater, K. F. & Horinouchi, S. (2003) Mol. Microbiol. 48, 9-15. - PubMed
-
- Willey, J. M., Santamaria, R., Guijarro, J., Geistlich, M. & Losick, R. (1991) Cell 65, 641-650. - PubMed
-
- Tillotson, R. D., Wösten, H. A. B., Richter, M. & Willey, J. M. (1998) Mol. Microbiol. 30, 595-602. - PubMed
-
- Chater, K. F. (2000) in Prokaryote Development, eds. Brun, Y. V. & Shimkets, L. J. (Am. Soc. Microbiol., Washington, DC), pp. 33-48.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

