RNA dynamics in live Escherichia coli cells
- PMID: 15277674
- PMCID: PMC509199
- DOI: 10.1073/pnas.0404443101
RNA dynamics in live Escherichia coli cells
Abstract
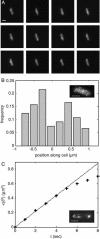
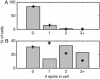
We describe a method for tracking RNA molecules in Escherichia coli that is sensitive to single copies of mRNA, and, using the method, we find that individual molecules can be followed for many hours in living cells. We observe distinct characteristic dynamics of RNA molecules, all consistent with the known life history of RNA in prokaryotes: localized motion consistent with the Brownian motion of an RNA polymer tethered to its template DNA, free diffusion, and a few examples of polymer chain dynamics that appear to be a combination of chain fluctuation and chain elongation attributable to RNA transcription. We also quantify some of the dynamics, such as width of the displacement distribution, diffusion coefficient, chain elongation rate, and distribution of molecule numbers, and compare them with known biophysical parameters of the E. coli system.
Figures



References
-
- Neidhardt, F. C., Ingraham, J. L. & Schaechter, M. (1990) Physiology of the Bacterial Cell: A Molecular Approach (Sinauer, Sunderland, MA).
-
- Neidhardt, F. C. (1996) Escherichia coli and Salmonella: Cellular and Molecular Biology (Am. Soc. Microbiol., Washington, DC).
-
- Errington, J. (2003) Nat. Cell Biol. 5, 175–178. - PubMed
-
- Southward, C. M. & Surette, M. G. (2002) Mol. Microbiol. 45, 1191–1196. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

