Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells
- PMID: 15277680
- PMCID: PMC509193
- DOI: 10.1073/pnas.0400541101
Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells
Abstract
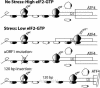
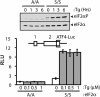
During cellular stresses, phosphorylation of eukaryotic initiation factor-2 (eIF2) elicits gene expression designed to ameliorate the underlying cellular disturbance. Central to this stress response is the transcriptional regulator activating transcription factor, ATF4. Here we describe the mechanism regulating ATF4 expression involving the differential contribution of two upstream ORFs (uORFs) in the 5' leader of the mouse ATF4 mRNA. The 5' proximal uORF1 is a positive-acting element that facilitates ribosome scanning and reinitiation at downstream coding regions in the ATF4 mRNA. When eIF2-GTP is abundant in nonstressed cells, ribosomes scanning downstream of uORF1 reinitiate at the next coding region, uORF2, an inhibitory element that blocks ATF4 expression. During stress conditions, phosphorylation of eIF2 and the accompanying reduction in the levels of eIF2-GTP increase the time required for the scanning ribosomes to become competent to reinitiate translation. This delayed reinitiation allows for ribosomes to scan through the inhibitory uORF2 and instead reinitiate at the ATF4-coding region. Increased expression of ATF4 would contribute to the expression of genes involved in remediation of cellular stress damage. These results suggest that the mechanism of translation reinitiation involving uORFs is conserved from yeast to mammals.
Figures



Similar articles
-
Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions.J Biol Chem. 2008 Mar 14;283(11):7064-73. doi: 10.1074/jbc.M708530200. Epub 2008 Jan 14. J Biol Chem. 2008. PMID: 18195013
-
Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response.J Cell Biol. 2004 Oct 11;167(1):27-33. doi: 10.1083/jcb.200408003. J Cell Biol. 2004. PMID: 15479734 Free PMC article.
-
Requirements for intercistronic distance and level of eukaryotic initiation factor 2 activity in reinitiation on GCN4 mRNA vary with the downstream cistron.Mol Cell Biol. 1994 Apr;14(4):2616-28. doi: 10.1128/mcb.14.4.2616-2628.1994. Mol Cell Biol. 1994. PMID: 8139562 Free PMC article.
-
Does eIF3 promote reinitiation after translation of short upstream ORFs also in mammalian cells?RNA Biol. 2017 Dec 2;14(12):1660-1667. doi: 10.1080/15476286.2017.1353863. Epub 2017 Sep 15. RNA Biol. 2017. PMID: 28745933 Free PMC article. Review.
-
Gene-specific translational control of the yeast GCN4 gene by phosphorylation of eukaryotic initiation factor 2.Mol Microbiol. 1993 Oct;10(2):215-23. doi: 10.1111/j.1365-2958.1993.tb01947.x. Mol Microbiol. 1993. PMID: 7934812 Review.
Cited by
-
Mammalian target of rapamycin complex 1 (mTORC1) enhances bortezomib-induced death in tuberous sclerosis complex (TSC)-null cells by a c-MYC-dependent induction of the unfolded protein response.J Biol Chem. 2013 May 31;288(22):15687-98. doi: 10.1074/jbc.M112.431056. Epub 2013 Apr 23. J Biol Chem. 2013. PMID: 23612979 Free PMC article.
-
Inhibition of the integrated stress response by viral proteins that block p-eIF2-eIF2B association.Nat Microbiol. 2020 Nov;5(11):1361-1373. doi: 10.1038/s41564-020-0759-0. Epub 2020 Jul 20. Nat Microbiol. 2020. PMID: 32690955
-
Translational Upregulation of an Individual p21Cip1 Transcript Variant by GCN2 Regulates Cell Proliferation and Survival under Nutrient Stress.PLoS Genet. 2015 Jun 23;11(6):e1005212. doi: 10.1371/journal.pgen.1005212. eCollection 2015 Jun. PLoS Genet. 2015. PMID: 26102367 Free PMC article.
-
Stress-induced C/EBP homology protein (CHOP) represses MyoD transcription to delay myoblast differentiation.PLoS One. 2011;6(12):e29498. doi: 10.1371/journal.pone.0029498. Epub 2011 Dec 29. PLoS One. 2011. PMID: 22242125 Free PMC article.
-
Physiological and pathophysiological role of nonsense-mediated mRNA decay.Pflugers Arch. 2016 Jun;468(6):1013-28. doi: 10.1007/s00424-016-1826-5. Epub 2016 Apr 30. Pflugers Arch. 2016. PMID: 27138169 Review.
References
-
- Hinnebusch, A. G. (1997) J. Biol. Chem. 272, 21661–21664. - PubMed
-
- Hinnebusch, A. G. (2000) in Translational Control of Gene Expression, eds. Sonenberg, N., Hershey, J. W. B. & Mathews, M. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 185–244.
-
- Wek, R. C. (1994) Trends Biochem. Sci. 19, 491–496. - PubMed
-
- Dever, T. E. (2002) Cell 108, 545–556. - PubMed
-
- Williams, B. R. (1999) Oncogene 18, 6112–6120. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

