Initial cleavage of the human immunodeficiency virus type 1 GagPol precursor by its activated protease occurs by an intramolecular mechanism
- PMID: 15280456
- PMCID: PMC479095
- DOI: 10.1128/JVI.78.16.8477-8485.2004
Initial cleavage of the human immunodeficiency virus type 1 GagPol precursor by its activated protease occurs by an intramolecular mechanism
Abstract
Processing of the GagPol polyprotein precursor of human immunodeficiency virus type 1 (HIV-1) is a critical step in viral assembly and replication. The HIV-1 protease (PR) is translated as part of GagPol and is both necessary and sufficient for precursor processing. The PR is active only as a dimer; enzyme activation is initiated when the PR domains in two GagPol precursors dimerize. The precise mechanism by which the PR becomes activated and the subsequent initial steps in precursor processing are not well understood. However, it is clear that processing is initiated by the PR domain that is embedded within the precursor itself. We have examined the earliest events in precursor processing using an in vitro assay in which full-length GagPol is cleaved by its embedded PR. We demonstrate that the embedded, immature PR is as much as 10,000-fold less sensitive to inhibition by an active-site PR inhibitor than is the mature, free enzyme. Further, we find that different concentrations of the active-site inhibitor are required to inhibit the processing of different cleavage sites within GagPol. Finally, our results indicate that the first cleavages carried out by the activated PR within GagPol are intramolecular. Overall, our data support a model of virus assembly in which the first cleavages occur in GagPol upstream of the PR. These intramolecular cleavages produce an extended form of PR that completes the final processing steps accompanying the final stages of particle assembly by an intermolecular mechanism.
Figures


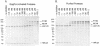
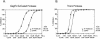
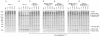
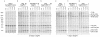
Similar articles
-
Ordered processing of the human immunodeficiency virus type 1 GagPol precursor is influenced by the context of the embedded viral protease.J Virol. 2005 Aug;79(16):10601-7. doi: 10.1128/JVI.79.16.10601-10607.2005. J Virol. 2005. PMID: 16051852 Free PMC article.
-
Processing sites in the human immunodeficiency virus type 1 (HIV-1) Gag-Pro-Pol precursor are cleaved by the viral protease at different rates.Retrovirology. 2005 Nov 1;2:66. doi: 10.1186/1742-4690-2-66. Retrovirology. 2005. PMID: 16262906 Free PMC article.
-
The dimer interfaces of protease and extra-protease domains influence the activation of protease and the specificity of GagPol cleavage.J Virol. 2003 Jan;77(1):366-74. doi: 10.1128/jvi.77.1.366-374.2003. J Virol. 2003. PMID: 12477841 Free PMC article.
-
Beyond Inhibition: A Novel Strategy of Targeting HIV-1 Protease to Eliminate Viral Reservoirs.Viruses. 2022 May 28;14(6):1179. doi: 10.3390/v14061179. Viruses. 2022. PMID: 35746649 Free PMC article. Review.
-
Cellular Targets of HIV-1 Protease: Just the Tip of the Iceberg?Viruses. 2023 Mar 9;15(3):712. doi: 10.3390/v15030712. Viruses. 2023. PMID: 36992421 Free PMC article. Review.
Cited by
-
Membrane-Active Sequences within gp41 Membrane Proximal External Region (MPER) Modulate MPER-Containing Peptidyl Fusion Inhibitor Activity and the Biosynthesis of HIV-1 Structural Proteins.PLoS One. 2015 Jul 31;10(7):e0134851. doi: 10.1371/journal.pone.0134851. eCollection 2015. PLoS One. 2015. PMID: 26230322 Free PMC article.
-
Autoprocessing of human immunodeficiency virus type 1 protease miniprecursor fusions in mammalian cells.AIDS Res Ther. 2010 Jul 28;7:27. doi: 10.1186/1742-6405-7-27. AIDS Res Ther. 2010. PMID: 20667109 Free PMC article.
-
Design and Synthesis of Potent HIV-1 Protease Inhibitors Containing Bicyclic Oxazolidinone Scaffold as the P2 Ligands: Structure-Activity Studies and Biological and X-ray Structural Studies.J Med Chem. 2018 Nov 8;61(21):9722-9737. doi: 10.1021/acs.jmedchem.8b01227. Epub 2018 Oct 24. J Med Chem. 2018. PMID: 30354121 Free PMC article.
-
Drug-associated changes in amino acid residues in Gag p2, p7(NC), and p6(Gag)/p6(Pol) in human immunodeficiency virus type 1 (HIV-1) display a dominant effect on replicative fitness and drug response.Virology. 2008 Sep 1;378(2):272-81. doi: 10.1016/j.virol.2008.05.029. Epub 2008 Jul 2. Virology. 2008. PMID: 18599104 Free PMC article.
-
Detection of Gag C-terminal mutations among HIV-1 non-B subtypes in a subset of Cameroonian patients.Sci Rep. 2022 Jan 26;12(1):1374. doi: 10.1038/s41598-022-05375-9. Sci Rep. 2022. PMID: 35082353 Free PMC article.
References
-
- Bruice, T. C., and S. Benkovic. 1966. Bioorganic mechanisms, p. 119-125. W. A. Benjamin, New York, N.Y.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

