Analysis of integration sites of Jaagsiekte sheep retrovirus in ovine pulmonary adenocarcinoma
- PMID: 15280459
- PMCID: PMC479065
- DOI: 10.1128/JVI.78.16.8506-8512.2004
Analysis of integration sites of Jaagsiekte sheep retrovirus in ovine pulmonary adenocarcinoma
Abstract
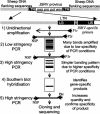
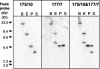
Ovine pulmonary adenocarcinoma (OPA) is an infectious lung tumor of sheep caused by Jaagsiekte sheep retrovirus (JSRV). To test the hypothesis that JSRV insertional mutagenesis is involved in the oncogenesis of OPA, we cloned and characterized 70 independent integration sites from 23 cases of OPA. Multiple integration sites were identified in most tumors. BLAST analysis of the sequences did not disclose any potential oncogenic motifs or any identical integration sites in different tumors. Thirty-seven of the integration sites were mapped to individual chromosomes by PCR with a panel of sheep-hamster hybrid cell lines. Integration sites were found on 20 of the 28 sheep chromosomes, suggesting a random distribution. However, four integration sites from four different tumors mapped to chromosome 16. By Southern blot hybridization, probes derived from two of these sites mapped to within 5 kb of each other on normal sheep DNA. These sites were found within a single sheep bacterial artificial chromosome clone and were further mapped to only 2.5 kb apart, within an uncharacterized predicted gene and less than 200 kb from a mitogen-activated protein kinase-encoding gene. These findings suggest that there is at least one common integration site for JSRV in OPA and add weight to the hypothesis that insertional mutagenesis is involved in the development of this tumor.
Figures


References
-
- Aguilar-Cordova, E., R. Strange, L. J., Young, H. T. Billy, P. H. Gumerlock, and R. D. Cardiff. 1991. Viral Ha-ras mediated mammary tumor progression. Oncogene 6:1601-1607. - PubMed
-
- Allen, T. E., K. J. Sherrill, S. M. Crispell, M. R. Perrott, J. O. Carlson, and J. C. DeMartini. 2002. The Jaagsiekte retrovirus envelope gene induces transformation of the avian embryo fibroblast cell line DF-1 but does not require a conserved SH2 binding domain. J. Gen. Virol. 83:2733-2742. - PubMed
-
- Brown, P. O. 1997. Integration, p. 106-203. In J. M. Coffin et al. (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
-
- Burkin, D. J., T. E. Broad, M. R. Lambeth, H. R. Burkin, and C. Jones. 1998. New gene assignments using a complete, characterized sheep-hamster somatic cell hybrid panel. Anim. Genet. 29:48-54. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

