Basal core promoter and precore mutations in the hepatitis B virus genome enhance replication efficacy of Lamivudine-resistant mutants
- PMID: 15280461
- PMCID: PMC479060
- DOI: 10.1128/JVI.78.16.8524-8535.2004
Basal core promoter and precore mutations in the hepatitis B virus genome enhance replication efficacy of Lamivudine-resistant mutants
Abstract
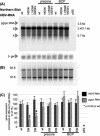
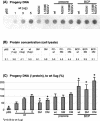
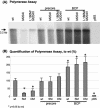
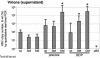
During chronic hepatitis B virus (HBV) infection, mutations in the precore (PC) or basal core promoter (BCP) region affecting HBV e antigen (HBeAg) expression occur commonly and represent the predominant virus species in patients with HBeAg-negative chronic hepatitis B. The PC mutation (G1896A+C1858T) creates a translational stop codon resulting in absent HBeAg expression, whereas BCP mutations (A1762T/G1764A) reduce HBeAg expression by transcriptional mechanisms. Treatment of chronic HBV infection with lamivudine (LMV) often selects drug-resistant strains with single (rtM204I) or double (rtL180M+rtM204V) point mutations in the YMDD motif of HBV reverse transcriptase. We cloned replication-competent HBV vectors (genotype A, adw2) combining mutations in the core (wild type [wt], PC, and BCP) and polymerase gene (wt, rtM204I, and rtL180M/M204V) and analyzed virus replication and drug sensitivity in vitro. Resistance to LMV (rtM204I/rtL180M+rtM204V) was accompanied by a reduced replication efficacy as evidenced by reduced pregenomic RNA, encapsidated progeny DNA, polymerase activity, and virion release. PC mutations alone did not alter virus replication but restored replication efficacy of the LMV-resistant mutants without affecting drug resistance. BCP mutants had higher replication capacities than did the wt, also in combination with LMV resistance mutations. All nine HBV constructs showed similar sensitivities to adefovir. In conclusion, BCP-PC mutations directly impact the replication capacity of LMV-resistant mutants. PC mutations compensated for replication inefficiency of LMV-resistant mutants, whereas BCP mutations increased viral replication levels to above the wt baseline values, even in LMV-resistant mutants, without affecting drug sensitivity in vitro. Adefovir may be an effective treatment when combinations of core and polymerase mutations occur.
Figures




Similar articles
-
Impact of hepatitis B e antigen-suppressing mutations on the replication efficiency of entecavir-resistant hepatitis B virus strains.J Viral Hepat. 2011 Nov;18(11):804-14. doi: 10.1111/j.1365-2893.2010.01378.x. Epub 2010 Sep 30. J Viral Hepat. 2011. PMID: 20887378
-
The YMDD and rtA194T mutations result in decreased replication capacity in wild-type HBV as well as in HBV with precore and basal core promoter mutations.Antivir Chem Chemother. 2011 Aug 23;22(1):13-22. doi: 10.3851/IMP1791. Antivir Chem Chemother. 2011. PMID: 21860069
-
Effect of the G1896A precore mutation on drug sensitivity and replication yield of lamivudine-resistant HBV in vitro.Hepatology. 2003 Jan;37(1):27-35. doi: 10.1053/jhep.2003.50012. Hepatology. 2003. PMID: 12500185
-
The virological and clinical significance of mutations in the overlapping envelope and polymerase genes of hepatitis B virus.J Clin Virol. 2002 Aug;25(2):97-106. doi: 10.1016/s1386-6532(02)00049-5. J Clin Virol. 2002. PMID: 12367644 Review.
-
Influence of mutations in the hepatitis B virus genome on virus replication and drug resistance--implications for novel antiviral strategies.Curr Med Chem. 2004 Oct;11(20):2667-77. doi: 10.2174/0929867043364333. Curr Med Chem. 2004. PMID: 15544468 Review.
Cited by
-
Epidemiology, Genotyping, Mutational and Phylogenetic Analysis of Hepatitis B Virus Infection in North-east India.J Clin Exp Hepatol. 2022 Jan-Feb;12(1):43-51. doi: 10.1016/j.jceh.2021.04.002. Epub 2021 Apr 19. J Clin Exp Hepatol. 2022. PMID: 35068784 Free PMC article.
-
Detection of hepatitis B virus A1762T/G1764A mutant by amplification refractory mutation system.Braz J Infect Dis. 2014 May-Jun;18(3):261-5. doi: 10.1016/j.bjid.2013.09.005. Epub 2014 Jan 3. Braz J Infect Dis. 2014. PMID: 24389280 Free PMC article.
-
Etiology and Viral Genotype in Patients with End-Stage Liver Diseases admitted to a Hepatology Unit in Colombia.Hepat Res Treat. 2011;2011:363205. doi: 10.1155/2011/363205. Epub 2011 Sep 20. Hepat Res Treat. 2011. PMID: 21941645 Free PMC article.
-
Analysis of HBV basal core promoter/precore gene variability in patients with HBV drug resistance and HIV co-infection in Northwest Ethiopia.PLoS One. 2018 Feb 6;13(2):e0191970. doi: 10.1371/journal.pone.0191970. eCollection 2018. PLoS One. 2018. PMID: 29408943 Free PMC article.
-
Age, race and viral genotype are associated with the prevalence of hepatitis B e antigen in children and adults with chronic hepatitis B.J Viral Hepat. 2019 Jul;26(7):856-865. doi: 10.1111/jvh.13104. Epub 2019 May 2. J Viral Hepat. 2019. PMID: 30974509 Free PMC article.
References
-
- Angus, P., R. Vaughan, S. Xiong, H. Yang, W. E. Delaney, C. Gibbs, C. L. Brosgart, D. Colledge, R. Edwards, A. Ayres, A. Bartholomeusz, and S. Locarnini. 2003. Resistance to adefovir dipivoxil therapy associated with the selection of a novel mutation in the HBV polymerase. Gastroenterology 125:292-297. - PubMed
-
- Baptista, M., A. Kramvis, and M. C. Kew. 1999. High prevalence of 1762(T) 1764(A) mutations in the basic core promoter of hepatitis B virus isolated from black Africans with hepatocellular carcinoma compared with asymptomatic carriers. Hepatology 29:946-953. - PubMed
-
- Bock, C. T., B. Buerke, H. L. Tillmann, F. Tacke, V. Kliem, M. P. Manns, and C. Trautwein. 2003. Relevance of hepatitis B core gene deletions in patients after kidney transplantation. Gastroenterology 124:1809-1820. - PubMed
-
- Bock, C. T., H. L. Tillmann, H. J. Maschek, M. P. Manns, and C. Trautwein. 1997. A preS mutation isolated from a patient with chronic hepatitis B infection leads to virus retention and misassembly. Gastroenterology 113:1976-1982. - PubMed
-
- Bock, C. T., H. L. Tillmann, J. Torresi, J. Klempnauer, S. Locarnini, M. P. Manns, and C. Trautwein. 2002. Selection of hepatitis B virus polymerase mutants with enhanced replication by lamivudine treatment after liver transplantation. Gastroenterology 122:264-273. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

