Characterization of nucleocapsid binding by the measles virus and mumps virus phosphoproteins
- PMID: 15280472
- PMCID: PMC479056
- DOI: 10.1128/JVI.78.16.8630-8640.2004
Characterization of nucleocapsid binding by the measles virus and mumps virus phosphoproteins
Erratum in
- J Virol. 2005 Aug;79(15):10097
Abstract
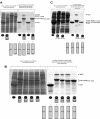
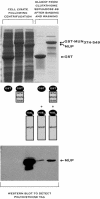
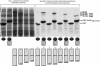
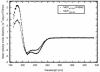
We report an analysis of the interaction between the P protein and the RNA-associated N protein (N-RNA) for both measles and mumps viruses with proteins produced in a bacterial expression system. During this study, we verified that the C-terminal tail of the N protein is not required for nucleocapsid formation. For both measles and mumps virus N, truncated proteins encompassing amino acids 1 to 375 assemble into nucleocapsid-like particles within the bacterial cell. For measles virus N, the binding site for the P protein maps to residues 477 to 505 within the tail of the molecule, a sequence relatively conserved among the morbilliviruses. For mumps virus N, a binding site for the P protein maps to the assembly domain of N (residues 1 to 398), while no strong binding of the P protein to the tail of N was detected. These results suggest that the site of attachment for the polymerase varies among the paramyxoviruses. Pulldown experiments demonstrate that the last 50 amino acids of both measles virus and mumps virus P (measles virus P, 457 to 507; mumps virus P, 343 to 391) by themselves constitute the nucleocapsid-binding domain (NBD). Spectroscopic studies show that the NBD is predominantly alpha-helical in both viruses. However, only in measles virus P is the NBD stable and folded, having a lesser degree of tertiary organization in mumps virus P. With isothermal titration calorimetry, we demonstrate that the measles virus P NBD binds to residues 477 to 505 of measles virus N with 1:1 stoichiometry. The dissociation constant (K(d)) was determined to be 13 microM at 20 degrees C and 35 microM at 37 degrees C. Our data are consistent with a model in which an alpha-helical nucleocapsid binding domain, located at the C terminus of P, is responsible for tethering the viral polymerase to its template yet also suggest that, in detail, polymerase binding in morbilliviruses and rubulaviruses differs significantly.
Figures



References
-
- Amexis, G., S. Rubin, V. Chizhikov, F. Pelloquin, K. Carbone, and K. Chumakov. 2002. Sequence diversity of Jeryl Lynn strain of mumps virus: quantitative mutant analysis for vaccine quality control. Virology 300:171-179. - PubMed
-
- Bankamp, B., S. M. Horikami, P. D. Thompson, M. Huber, M. Billeter, and S. A. Moyer. 1996. Domains of the measles virus N protein required for binding to P protein and self-assembly. Virology 216:272-277. - PubMed
-
- Bhella, D., A. Ralph, L. B. Murphy, and R. P. Yeo. 2002. Significant differences in nucleocapsid morphology within the Paramyxoviridae. J. Gen. Virol. 83:1831-1839. - PubMed
-
- Blixenkrone-Moller, M., G. Bolt, E. Gottschalck, and M. Kenter. 1994. Comparative analysis of the gene encoding the nucleocapsid protein of dolphin morbillivirus reveals its distant evolutionary relationship to measles virus and ruminant morbilliviruses. J. Gen. Virol. 75:2829-2834. - PubMed
-
- Buchholz, C. J., C. Retzler, H. E. Homann, and W. J. Neubert. 1994. The carboxy-terminal domain of Sendai virus nucleocapsid protein is involved in complex formation between phosphoprotein and nucleocapsid-like particles. Virology 204:770-776. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

