Antiretroviral drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase increase template-switching frequency
- PMID: 15280484
- PMCID: PMC479068
- DOI: 10.1128/JVI.78.16.8761-8770.2004
Antiretroviral drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase increase template-switching frequency
Abstract
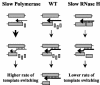
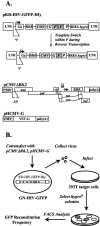
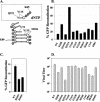
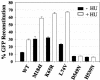
Template-switching events during reverse transcription are necessary for completion of retroviral replication and recombination. Structural determinants of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) that influence its template-switching frequency are not known. To identify determinants of HIV-1 RT that affect the frequency of template switching, we developed an in vivo assay in which RT template-switching events during viral replication resulted in functional reconstitution of the green fluorescent protein gene. A survey of single amino acid substitutions near the polymerase active site or deoxynucleoside triphosphate-binding site of HIV-1 RT indicated that several substitutions increased the rate of RT template switching. Several mutations associated with resistance to antiviral nucleoside analogs (K65R, L74V, E89G, Q151N, and M184I) dramatically increased RT template-switching frequencies by two- to sixfold in a single replication cycle. In contrast, substitutions in the RNase H domain (H539N, D549N) decreased the frequency of RT template switching by twofold. Depletion of intracellular nucleotide pools by hydroxyurea treatment of cells used as targets for infection resulted in a 1.8-fold increase in the frequency of RT template switching. These results indicate that the dynamic steady state between polymerase and RNase H activities is an important determinant of HIV-1 RT template switching and establish that HIV-1 recombination occurs by the previously described dynamic copy choice mechanism. These results also indicate that mutations conferring resistance to antiviral drugs can increase the frequency of RT template switching and may influence the rate of retroviral recombination and viral evolution.
Figures

References
-
- An, W. F., and A. Telesnitsky. 2001. Frequency of direct repeat deletion in a human immunodeficiency virus type 1 vector during reverse transcription in human cells. Virology 286:475-482. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

