Budding yeast silencing complexes and regulation of Sir2 activity by protein-protein interactions
- PMID: 15282295
- PMCID: PMC479720
- DOI: 10.1128/MCB.24.16.6931-6946.2004
Budding yeast silencing complexes and regulation of Sir2 activity by protein-protein interactions
Abstract
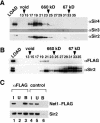
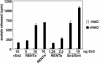
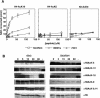
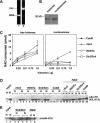
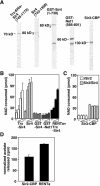
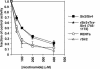
Gene silencing in the budding yeast Saccharomyces cerevisiae requires the enzymatic activity of the Sir2 protein, a highly conserved NAD-dependent deacetylase. In order to study the activity of native Sir2, we purified and characterized two budding yeast Sir2 complexes: the Sir2/Sir4 complex, which mediates silencing at mating-type loci and at telomeres, and the RENT complex, which mediates silencing at the ribosomal DNA repeats. Analyses of the protein compositions of these complexes confirmed previously described interactions. We show that the assembly of Sir2 into native silencing complexes does not alter its selectivity for acetylated substrates, nor does it allow the deacetylation of nucleosomal histones. The inability of Sir2 complexes to deacetylate nucleosomes suggests that additional factors influence Sir2 activity in vivo. In contrast, Sir2 complexes show significant enhancement in their affinities for acetylated substrates and their sensitivities to the physiological inhibitor nicotinamide relative to recombinant Sir2. Reconstitution experiments showed that, for the Sir2/Sir4 complex, these differences stem from the physical interaction of Sir2 with Sir4. Finally, we provide evidence that the different nicotinamide sensitivities of Sir2/Sir4 and RENT in vitro could contribute to locus-specific differences in how Sir2 activity is regulated in vivo.
Figures



References
-
- Avalos, J. L., I. Celic, S. Muhammad, M. S. Cosgrove, J. D. Boeke, and C. Wolberger. 2002. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol. Cell 10:523-535. - PubMed
-
- Bell, S. D., C. H. Botting, B. N. Wardleworth, S. P. Jackson, and M. F. White. 2002. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296:148-151. - PubMed
-
- Bitterman, K. J., R. M. Anderson, H. Y. Cohen, M. Latorre-Esteves, and D. A. Sinclair. 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 277:45099-45107. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
