RPAP1, a novel human RNA polymerase II-associated protein affinity purified with recombinant wild-type and mutated polymerase subunits
- PMID: 15282305
- PMCID: PMC479746
- DOI: 10.1128/MCB.24.16.7043-7058.2004
RPAP1, a novel human RNA polymerase II-associated protein affinity purified with recombinant wild-type and mutated polymerase subunits
Abstract
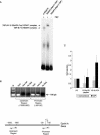
We have programmed human cells to express physiological levels of recombinant RNA polymerase II (RNAPII) subunits carrying tandem affinity purification (TAP) tags. Double-affinity chromatography allowed for the simple and efficient isolation of a complex containing all 12 RNAPII subunits, the general transcription factors TFIIB and TFIIF, the RNAPII phosphatase Fcp1, and a novel 153-kDa polypeptide of unknown function that we named RNAPII-associated protein 1 (RPAP1). The TAP-tagged RNAPII complex is functionally active both in vitro and in vivo. A role for RPAP1 in RNAPII transcription was established by shutting off the synthesis of Ydr527wp, a Saccharomyces cerevisiae protein homologous to RPAP1, and demonstrating that changes in global gene expression were similar to those caused by the loss of the yeast RNAPII subunit Rpb11. We also used TAP-tagged Rpb2 with mutations in fork loop 1 and switch 3, two structural elements located strategically within the active center, to start addressing the roles of these elements in the interaction of the enzyme with the template DNA during the transcription reaction.
Figures




References
-
- Acker, J., M. Wintzerith, M. Vigneron, and C. Kedinger. 1992. Primary structure of the second largest subunit of human RNA polymerase II (or B). J. Mol. Biol. 226:1295-1299. - PubMed
-
- Awrey, D. E., R. G. Weilbaecher, S. A. Hemming, S. M. Orlicky, C. M. Kane, and A. M. Edwards. 1997. Transcription elongation through DNA arrest sites. A multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. J. Biol. Chem. 272:14747-14754. - PubMed
-
- Bagby, S., S. Kim, E. Maldonado, K. I. Tong, D. Reinberg, and M. Ikura. 1995. Solution structure of the C-terminal core domain of human TFIIB: similarity to cyclin A and interaction with TATA-binding protein. Cell 82:857-867. - PubMed
-
- Bryant, G. O., and M. Ptashne. 2003. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol. Cell 11:1301-1309. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
