Scrambled prion domains form prions and amyloid
- PMID: 15282319
- PMCID: PMC479727
- DOI: 10.1128/MCB.24.16.7206-7213.2004
Scrambled prion domains form prions and amyloid
Abstract
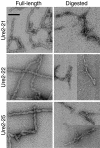
The [URE3] prion of Saccharomyces cerevisiae is a self-propagating amyloid form of Ure2p. The amino-terminal prion domain of Ure2p is necessary and sufficient for prion formation and has a high glutamine (Q) and asparagine (N) content. Such Q/N-rich domains are found in two other yeast prion proteins, Sup35p and Rnq1p, although none of the many other yeast Q/N-rich domain proteins have yet been found to be prions. To examine the role of amino acid sequence composition in prion formation, we used Ure2p as a model system and generated five Ure2p variants in which the order of the amino acids in the prion domain was randomly shuffled while keeping the amino acid composition and C-terminal domain unchanged. Surprisingly, all five formed prions in vivo, with a range of frequencies and stabilities, and the prion domains of all five readily formed amyloid fibers in vitro. Although it is unclear whether other amyloid-forming proteins would be equally resistant to scrambling, this result demonstrates that [URE3] formation is driven primarily by amino acid composition, largely independent of primary sequence.
Figures




Similar articles
-
Primary sequence independence for prion formation.Proc Natl Acad Sci U S A. 2005 Sep 6;102(36):12825-30. doi: 10.1073/pnas.0506136102. Epub 2005 Aug 25. Proc Natl Acad Sci U S A. 2005. PMID: 16123127 Free PMC article.
-
A promiscuous prion: efficient induction of [URE3] prion formation by heterologous prion domains.Genetics. 2009 Nov;183(3):929-40. doi: 10.1534/genetics.109.109322. Epub 2009 Sep 14. Genetics. 2009. PMID: 19752212 Free PMC article.
-
Yeast prions: evolution of the prion concept.Prion. 2007 Apr-Jun;1(2):94-100. doi: 10.4161/pri.1.2.4664. Epub 2007 Apr 28. Prion. 2007. PMID: 19164928 Free PMC article. Review.
-
Prion amyloid structure explains templating: how proteins can be genes.FEMS Yeast Res. 2010 Dec;10(8):980-91. doi: 10.1111/j.1567-1364.2010.00666.x. FEMS Yeast Res. 2010. PMID: 20726897 Free PMC article. Review.
-
Amyloids of shuffled prion domains that form prions have a parallel in-register beta-sheet structure.Biochemistry. 2008 Apr 1;47(13):4000-7. doi: 10.1021/bi7024589. Epub 2008 Mar 7. Biochemistry. 2008. PMID: 18324784
Cited by
-
De novo design of synthetic prion domains.Proc Natl Acad Sci U S A. 2012 Apr 24;109(17):6519-24. doi: 10.1073/pnas.1119366109. Epub 2012 Apr 2. Proc Natl Acad Sci U S A. 2012. PMID: 22474356 Free PMC article.
-
Anti-prion systems in yeast.J Biol Chem. 2019 Feb 1;294(5):1729-1738. doi: 10.1074/jbc.TM118.004168. J Biol Chem. 2019. PMID: 30710020 Free PMC article. Review.
-
Determinants of histone H4 N-terminal domain function during nucleosomal array oligomerization: roles of amino acid sequence, domain length, and charge density.J Biol Chem. 2009 Jun 19;284(25):16716-16722. doi: 10.1074/jbc.M109.011288. Epub 2009 Apr 24. J Biol Chem. 2009. PMID: 19395382 Free PMC article.
-
Energy landscape of amyloidogenic peptide oligomerization by parallel-tempering molecular dynamics simulation: significant role of Asn ladder.Proc Natl Acad Sci U S A. 2005 Jun 7;102(23):8174-9. doi: 10.1073/pnas.0408653102. Epub 2005 May 27. Proc Natl Acad Sci U S A. 2005. PMID: 15923262 Free PMC article.
-
Genetic and epigenetic control of the efficiency and fidelity of cross-species prion transmission.Mol Microbiol. 2010 Jun;76(6):1483-99. doi: 10.1111/j.1365-2958.2010.07177.x. Epub 2010 Apr 23. Mol Microbiol. 2010. PMID: 20444092 Free PMC article.
References
-
- Bauer, H. H., U. Aebi, M. Haner, R. Hermann, M. Muller, and H. P. Merkle. 1995. Architecture and polymorphism of fibrillar supramolecular assemblies produced by in vitro aggregation of human calcitonin. J. Struct. Biol. 115:1-15. - PubMed
-
- Baxa, U., K. L. Taylor, J. S. Wall, M. N. Simon, N. Cheng, R. B. Wickner, and A. C. Steven. 2003. Architecture of Ure2p prion filaments: the N-terminal domains form a central core fiber. J. Biol. Chem. 278:43717-43727. - PubMed
-
- Bousset, L., H. Belrhali, J. Janin, R. Melki, and S. Morera. 2001. Structure of the globular region of the prion protein Ure2 from the yeast Saccharomyces cerevisiae. Structure 9:39-46. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
