Estrogens and progesterone promote persistent CCND1 gene activation during G1 by inducing transcriptional derepression via c-Jun/c-Fos/estrogen receptor (progesterone receptor) complex assembly to a distal regulatory element and recruitment of cyclin D1 to its own gene promoter
- PMID: 15282324
- PMCID: PMC479712
- DOI: 10.1128/MCB.24.16.7260-7274.2004
Estrogens and progesterone promote persistent CCND1 gene activation during G1 by inducing transcriptional derepression via c-Jun/c-Fos/estrogen receptor (progesterone receptor) complex assembly to a distal regulatory element and recruitment of cyclin D1 to its own gene promoter
Abstract
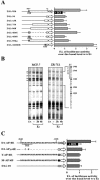
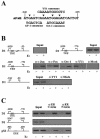
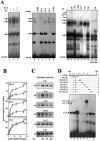
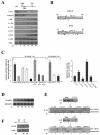
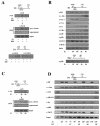
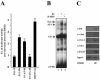
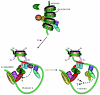
Transcriptional activation of the cyclin D1 gene (CCND1) plays a pivotal role in G(1)-phase progression, which is thereby controlled by multiple regulatory factors, including nuclear receptors (NRs). Appropriate CCND1 gene activity is essential for normal development and physiology of the mammary gland, where it is regulated by ovarian steroids through a mechanism(s) that is not fully elucidated. We report here that CCND1 promoter activation by estrogens in human breast cancer cells is mediated by recruitment of a c-Jun/c-Fos/estrogen receptor alpha complex to the tetradecanoyl phorbol acetate-responsive element of the gene, together with Oct-1 to a site immediately adjacent. This process coincides with the release from the same DNA region of a transcriptional repressor complex including Yin-Yang 1 (YY1) and histone deacetylase 1 and is sufficient to induce the assembly of the basal transcription machinery on the promoter and to lead to initial cyclin D1 accumulation in the cell. Later on in estrogen stimulation, the cyclin D1/Cdk4 holoenzyme associates with the CCND1 promoter, where E2F and pRb can also be found, contributing to the long-lasting gene enhancement required to drive G(1)-phase completion. Interestingly, progesterone triggers similar regulatory events through its own NRs, suggesting that the gene regulation cascade described here represents a crossroad for the transcriptional control of G(1)-phase progression by different classes of NRs.
Figures
Similar articles
-
Estrogen induction of the cyclin D1 promoter: involvement of a cAMP response-like element.Proc Natl Acad Sci U S A. 1999 Sep 28;96(20):11217-22. doi: 10.1073/pnas.96.20.11217. Proc Natl Acad Sci U S A. 1999. PMID: 10500157 Free PMC article.
-
Involvement of G1/S cyclins in estrogen-independent proliferation of estrogen receptor-positive breast cancer cells.Oncogene. 2002 Nov 21;21(53):8158-65. doi: 10.1038/sj.onc.1206012. Oncogene. 2002. PMID: 12444551
-
Fos and Jun inhibit estrogen-induced transcription of the human progesterone receptor gene through an activator protein-1 site.Mol Endocrinol. 2004 Mar;18(3):521-32. doi: 10.1210/me.2003-0105. Epub 2003 Dec 18. Mol Endocrinol. 2004. PMID: 14684847
-
Cyclin D1 determines estrogen signaling in the mammary gland in vivo.Mol Endocrinol. 2013 Sep;27(9):1415-28. doi: 10.1210/me.2013-1065. Epub 2013 Jul 17. Mol Endocrinol. 2013. PMID: 23864650 Free PMC article.
-
Prognostic and Predictive Value of CCND1/Cyclin D1 Amplification in Breast Cancer With a Focus on Postmenopausal Patients: A Systematic Review and Meta-Analysis.Front Endocrinol (Lausanne). 2022 Jun 17;13:895729. doi: 10.3389/fendo.2022.895729. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35784572 Free PMC article.
Cited by
-
Induction of cell proliferation and survival genes by estradiol-repressed microRNAs in breast cancer cells.BMC Cancer. 2012 Jan 20;12:29. doi: 10.1186/1471-2407-12-29. BMC Cancer. 2012. PMID: 22260523 Free PMC article.
-
Molecular determinants of context-dependent progesterone receptor action in breast cancer.BMC Med. 2014 Feb 20;12:32. doi: 10.1186/1741-7015-12-32. BMC Med. 2014. PMID: 24552158 Free PMC article. Review.
-
Estrogen receptor α-coupled Bmi1 regulation pathway in breast cancer and its clinical implications.BMC Cancer. 2014 Feb 24;14:122. doi: 10.1186/1471-2407-14-122. BMC Cancer. 2014. PMID: 24559156 Free PMC article.
-
1,1-dichloro-2,2-bis(p-chlorophenyl)ethylene (p,p'-DDE) disrupts the estrogen-androgen balance regulating the growth of hormone-dependent breast cancer cells.Breast Cancer Res. 2008;10(1):R16. doi: 10.1186/bcr1862. Epub 2008 Feb 14. Breast Cancer Res. 2008. PMID: 18275596 Free PMC article.
-
Co-treatment of mouse antral follicles with 17β-estradiol interferes with mono-2-ethylhexyl phthalate (MEHP)-induced atresia and altered apoptosis gene expression.Reprod Toxicol. 2014 Jun;45:45-51. doi: 10.1016/j.reprotox.2014.01.002. Epub 2014 Jan 9. Reprod Toxicol. 2014. PMID: 24412242 Free PMC article.
References
-
- Abbondanza, C., N. Medici, V. Nigro, V. Rossi, L. Gallo, G. Piluso, A. Belsito, A. Roscigno, P. Bontempo, A. A. Puca, A. M. Molinari, B. Moncharmont, and G. A. Puca. 2000. The retinoblastoma-interacting zinc-finger protein RIZ is a downstream effector of estrogen action. Proc. Natl. Acad. Sci. USA 97:3130-3135. - PMC - PubMed
-
- Adams, P. D. 2001. Regulation of the retinoblastoma tumor suppressor protein by cyclin cdks. Biochim. Biophys. Acta 1471:123-133. - PubMed
-
- Albanese, C., M. D'Amico, A. T. Reutens, M. Fu, G. Watanabe, R. J. Lee, R. N. Kitsis, B. Henglein, M. Avantaggiati, K. Somasundaram, B. Thimmapaya, and R. G. Pestell. 1999. Activation of the cyclin D1 gene by the E1A-associated protein p300 through AP-1 inhibits cellular apoptosis. J. Biol. Chem. 274:34186-34195. - PubMed
-
- Altucci, L., R. Addeo, L. Cicatiello, S. Dauvois, M. G. Parker, M. Truss, M. Beato, V. Sica, F. Bresciani, and A. Weisz. 1996. 17β-Estradiol induces cyclin D1 gene transcription, p36D1-p34cdk4 complex activation and p105Rb phosphorylation during mitogenic stimulation of G1-arrested human breast cancer cells. Oncogene 12:2315-2324. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
