EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3
- PMID: 15282325
- PMCID: PMC479730
- DOI: 10.1128/MCB.24.16.7275-7283.2004
EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3
Abstract
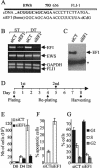
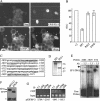
Ewing tumors are characterized by abnormal transcription factors resulting from the oncogenic fusion of EWS with members of the ETS family, most commonly FLI-1. RNA interference targeted to the junction between EWS and FLI-1 sequences was used to inactivate the EWS/FLI-1 fusion gene in Ewing cells and to explore the resulting phenotype and alteration of the gene expression profile. Loss of expression of EWS/FLI-1 resulted in the complete arrest of growth and was associated with a dramatic increase in the number of apoptotic cells. Gene profiling of Ewing cells in which the EWS/FLI-1 fusion gene had been inactivated identified downstream targets which could be grouped in two major functional clusters related to extracellular matrix structure or remodeling and regulation of signal transduction pathways. Among these targets, the insulin-like growth factor binding protein 3 gene (IGFBP-3), a major regulator of insulin-like growth factor 1 (IGF-1) proliferation and survival signaling, was strongly induced upon treating Ewing cells with EWS/FLI-1-specific small interfering RNAs. We show that EWS/FLI-1 can bind the IGFBP-3 promoter in vitro and in vivo and can repress its activity. Moreover, IGFBP-3 silencing can partially rescue the apoptotic phenotype caused by EWS/FLI-1 inactivation. Finally, IGFBP-3-induced Ewing cell apoptosis relies on both IGF-1-dependent and -independent pathways. These findings therefore identify the repression of IGFBP-3 as a key event in the development of Ewing's sarcoma.
Figures

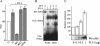
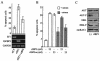
References
-
- Arvand, A., and C. T. Denny. 2001. Biology of EWS/ETS fusions in Ewing's family tumors. Oncogene 20:5747-5754. - PubMed
-
- Bailly, R. A., R. Bosselut, J. Zucman, F. Cormier, O. Delattre, M. Roussel, G. Thomas, and J. Ghysdael. 1994. DNA-binding and transcriptional activation properties of the EWS-FLI-1 fusion protein resulting from the t(11;22) translocation in Ewing sarcoma. Mol. Cell. Biol. 14:3230-3241. - PMC - PubMed
-
- Baserga, R. 1995. The insulin-like growth factor I receptor: a key to tumor growth? Cancer Res. 55:249-252. - PubMed
-
- Cohen, P., D. R. Clemmons, and R. G. Rosenfeld. 2000. Does the GH-IGF axis play a role in cancer pathogenesis? Growth Horm. IGF Res. 10:297-305. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
