A plant snoRNP complex containing snoRNAs, fibrillarin, and nucleolin-like proteins is competent for both rRNA gene binding and pre-rRNA processing in vitro
- PMID: 15282326
- PMCID: PMC479724
- DOI: 10.1128/MCB.24.16.7284-7297.2004
A plant snoRNP complex containing snoRNAs, fibrillarin, and nucleolin-like proteins is competent for both rRNA gene binding and pre-rRNA processing in vitro
Abstract
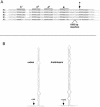
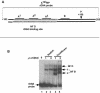
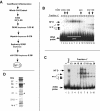
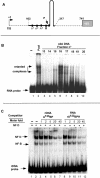
In eukaryotes the primary cleavage of the precursor rRNA (pre-rRNA) occurs in the 5' external transcribed spacer (5'ETS). In Saccharomyces cerevisiae and animals this cleavage depends on a conserved U3 small nucleolar ribonucleoprotein particle (snoRNP), including fibrillarin, and on other transiently associated proteins such as nucleolin. This large complex can be visualized by electron microscopy bound to the nascent pre-rRNA soon after initiation of transcription. Our group previously described a radish rRNA gene binding activity, NF D, that specifically binds to a cluster of conserved motifs preceding the primary cleavage site in the 5'ETS of crucifer plants including radish, cauliflower, and Arabidopsis thaliana (D. Caparros-Ruiz, S. Lahmy, S. Piersanti, and M. Echeverria, Eur. J. Biochem. 247:981-989, 1997). Here we report the purification and functional characterization of NF D from cauliflower inflorescences. Remarkably NF D also binds to 5'ETS RNA and accurately cleaves it at the primary cleavage site mapped in vivo. NF D is a multiprotein factor of 600 kDa that dissociates into smaller complexes. Two polypeptides of NF D identified by microsequencing are homologues of nucleolin and fibrillarin. The conserved U3 and U14 snoRNAs associated with fibrillarin and required for early pre-rRNA cleavages are also found in NF D. Based on this it is proposed that NF D is a processing complex that assembles on the rDNA prior to its interaction with the nascent pre-rRNA.
Figures

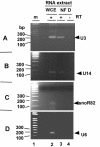
References
-
- Alba, M. M., and M. Pages. 1998. Plant proteins containing the RNA-recognition motif. Trends Plant Sci. 3:15-20.
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Barneche, F., F. Steinmetz, and M. Echeverria. 2000. Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J. Biol. Chem. 275:27212-27220. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
