Protein-protein interactions governing septin heteropentamer assembly and septin filament organization in Saccharomyces cerevisiae
- PMID: 15282341
- PMCID: PMC519150
- DOI: 10.1091/mbc.e04-04-0330
Protein-protein interactions governing septin heteropentamer assembly and septin filament organization in Saccharomyces cerevisiae
Abstract
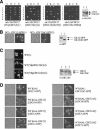
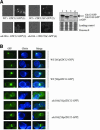
Mitotic yeast (Saccharomyces cerevisiae) cells express five related septins (Cdc3, Cdc10, Cdc11, Cdc12, and Shs1) that form a cortical filamentous collar at the mother-bud neck necessary for normal morphogenesis and cytokinesis. All five possess an N-terminal GTPase domain and, except for Cdc10, a C-terminal extension (CTE) containing a predicted coiled coil. Here, we show that the CTEs of Cdc3 and Cdc12 are essential for their association and for the function of both septins in vivo. Cdc10 interacts with a Cdc3-Cdc12 complex independently of the CTE of either protein. In contrast to Cdc3 and Cdc12, the Cdc11 CTE, which recruits the nonessential septin Shs1, is dispensable for its function in vivo. In addition, Cdc11 forms a stoichiometric complex with Cdc12, independent of its CTE. Reconstitution of various multiseptin complexes and electron microscopic analysis reveal that Cdc3, Cdc11, and Cdc12 are all necessary and sufficient for septin filament formation, and presence of Cdc10 causes filament pairing. These data provide novel insights about the connectivity among the five individual septins in functional septin heteropentamers and the organization of septin filaments.
Figures







Similar articles
-
Comprehensive Genetic Analysis of Paralogous Terminal Septin Subunits Shs1 and Cdc11 in Saccharomyces cerevisiae.Genetics. 2015 Jul;200(3):821-41. doi: 10.1534/genetics.115.176495. Epub 2015 May 12. Genetics. 2015. PMID: 25971665 Free PMC article.
-
Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4.J Cell Biol. 2004 Mar 1;164(5):701-15. doi: 10.1083/jcb.200312070. J Cell Biol. 2004. PMID: 14993234 Free PMC article.
-
Guanidine hydrochloride reactivates an ancient septin hetero-oligomer assembly pathway in budding yeast.Elife. 2020 Jan 28;9:e54355. doi: 10.7554/eLife.54355. Elife. 2020. PMID: 31990274 Free PMC article.
-
Septin mutations and phenotypes in S. cerevisiae.Cytoskeleton (Hoboken). 2019 Jan;76(1):33-44. doi: 10.1002/cm.21492. Epub 2018 Nov 18. Cytoskeleton (Hoboken). 2019. PMID: 30171672 Review.
-
Reuse, replace, recycle. Specificity in subunit inheritance and assembly of higher-order septin structures during mitotic and meiotic division in budding yeast.Cell Cycle. 2009 Jan 15;8(2):195-203. doi: 10.4161/cc.8.2.7381. Cell Cycle. 2009. PMID: 19164941 Free PMC article. Review.
Cited by
-
Bni5 tethers myosin-II to septins to enhance retrograde actin flow and the robustness of cytokinesis.bioRxiv [Preprint]. 2023 Nov 8:2023.11.07.566094. doi: 10.1101/2023.11.07.566094. bioRxiv. 2023. PMID: 37986946 Free PMC article. Preprint.
-
Actin and septin ultrastructures at the budding yeast cell cortex.Mol Biol Cell. 2005 Jan;16(1):372-84. doi: 10.1091/mbc.e04-08-0734. Epub 2004 Nov 3. Mol Biol Cell. 2005. PMID: 15525671 Free PMC article.
-
A Förster Resonance Energy Transfer (FRET)-based System Provides Insight into the Ordered Assembly of Yeast Septin Hetero-octamers.J Biol Chem. 2015 Nov 20;290(47):28388-28401. doi: 10.1074/jbc.M115.683128. Epub 2015 Sep 28. J Biol Chem. 2015. PMID: 26416886 Free PMC article.
-
Blastomyces dermatitidis septins CDC3, CDC10, and CDC12 impact the morphology of yeast and hyphae, but are not required for the phase transition.Med Mycol. 2013 Jan;51(1):93-102. doi: 10.3109/13693786.2012.699685. Epub 2012 Jul 12. Med Mycol. 2013. PMID: 22783804 Free PMC article.
-
Septin filament formation is essential in budding yeast.Dev Cell. 2011 Apr 19;20(4):540-9. doi: 10.1016/j.devcel.2011.02.004. Dev Cell. 2011. PMID: 21497764 Free PMC article.
References
-
- Barral, Y., Mermali, V., Mooseker, M.S., and Snyder, M. (2000). Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell 5, 841-851. - PubMed
-
- Beites, C.L., Xie, H., Bowser, R., and Trimble, W.S. (1999). The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat. Neurosci. 2, 434-439. - PubMed
-
- Brachmann, C.B., Davies, A., Cost, G.J., Caputo, E., Li, J., Hieter, P., and Boeke, J.D. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115-132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

