Genome-wide molecular dissection of serotype M3 group A Streptococcus strains causing two epidemics of invasive infections
- PMID: 15282372
- PMCID: PMC511060
- DOI: 10.1073/pnas.0404163101
Genome-wide molecular dissection of serotype M3 group A Streptococcus strains causing two epidemics of invasive infections
Abstract
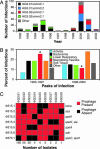
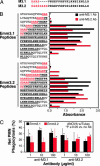
Molecular factors that contribute to the emergence of new virulent bacterial subclones and epidemics are poorly understood. We hypothesized that analysis of a population-based strain sample of serotype M3 group A Streptococcus (GAS) recovered from patients with invasive infection by using genome-wide investigative methods would provide new insight into this fundamental infectious disease problem. Serotype M3 GAS strains (n = 255) cultured from patients in Ontario, Canada, over 11 years and representing two distinct infection peaks were studied. Genetic diversity was indexed by pulsed-field gel electrophoresis, DNA-DNA microarray, whole-genome PCR scanning, prophage genotyping, targeted gene sequencing, and single-nucleotide polymorphism genotyping. All variation in gene content was attributable to acquisition or loss of prophages, a molecular process that generated unique combinations of proven or putative virulence genes. Distinct serotype M3 genotypes experienced rapid population expansion and caused infections that differed significantly in character and severity. Molecular genetic analysis, combined with immunologic studies, implicated a 4-aa duplication in the extreme N terminus of M protein as a factor contributing to an epidemic wave of serotype M3 invasive infections. This finding has implications for GAS vaccine research. Genome-wide analysis of population-based strain samples cultured from clinically well defined patients is crucial for understanding the molecular events underlying bacterial epidemics.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

