Visualization of RNA-protein interactions in living cells: FMRP and IMP1 interact on mRNAs
- PMID: 15282548
- PMCID: PMC514514
- DOI: 10.1038/sj.emboj.7600341
Visualization of RNA-protein interactions in living cells: FMRP and IMP1 interact on mRNAs
Abstract
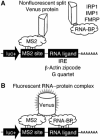
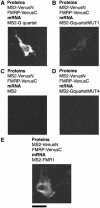
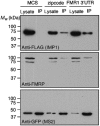
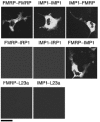
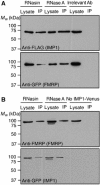
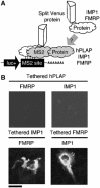
Protein expression depends significantly on the stability, translation efficiency and localization of mRNA. These qualities are largely dictated by the RNA-binding proteins associated with an mRNA. Here, we report a method to visualize and localize RNA-protein interactions in living mammalian cells. Using this method, we found that the fragile X mental retardation protein (FMRP) isoform 18 and the human zipcode-binding protein 1 ortholog IMP1, an RNA transport factor, were present on common mRNAs. These interactions occurred predominantly in the cytoplasm, in granular structures. In addition, FMRP and IMP1 interacted independently of RNA. Tethering of FMRP to an mRNA caused IMP1 to be recruited to the same mRNA and resulted in granule formation. The intimate association of FMRP and IMP1 suggests a link between mRNA transport and translational repression in mammalian cells.
Figures




References
-
- Antar LN, Bassell GJ (2003) Sunrise at the synapse: the FMRP mRNP shaping the synaptic interface. Neuron 37: 555–558 - PubMed
-
- Arn EA, Cha BJ, Theurkauf WE, Macdonald PM (2003) Recognition of a bicoid mRNA localization signal by a protein complex containing Swallow, Nod, and RNA binding proteins. Dev Cell 4: 41–51 - PubMed
-
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST (2001) Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107: 477–487 - PubMed
-
- Brown V, Small K, Lakkis L, Feng Y, Gunter C, Wilkinson KD, Warren ST (1998) Purified recombinant Fmrp exhibits selective RNA binding as an intrinsic property of the fragile X mental retardation protein. J Biol Chem 273: 15521–15527 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

